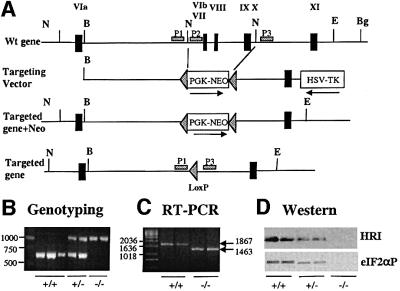
Fig. 1. Targeted disruption of the HRI gene. (A) HRI wild-type locus (top), targeting construct (middle) and targeted homologous recombination at the HRI locus before and after Cre-mediated excision of the neomycin resistance gene (bottom). Exons were marked by the filled rectangles and labeled with Roman numerals. Abbreviations used for restriction enzymes: N, NdeI; B, BssHII; E, EcoRV; Bg, BglII. (B) Genotyping of the targeted disrupted mice by PCR. The primers P1 and P2 were used for amplification of the HRI+/+ DNA of 625 bp, whereas primers P1 and P3 were used for the amplification of HRI–/– DNA of ∼1000 bp. (C) HRI mRNA. HRI mRNA was determined by RT–PCR from RNA in E19.5 fetal livers of HRI +/+ and –/– embryos. (D) HRI protein and eIF2α kinase activity. The levels of HRI protein in the lysates of 1 × 106 reticulocytes of HRI +/+, +/– and –/– mice were examined by western blot analysis (top). The eIF2α kinase activity of HRI in reticulocyte lysates was determined using in vitro protein kinase assays followed by western blot analysis using antibody specific to the phosphorylated eIF2α (bottom).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.