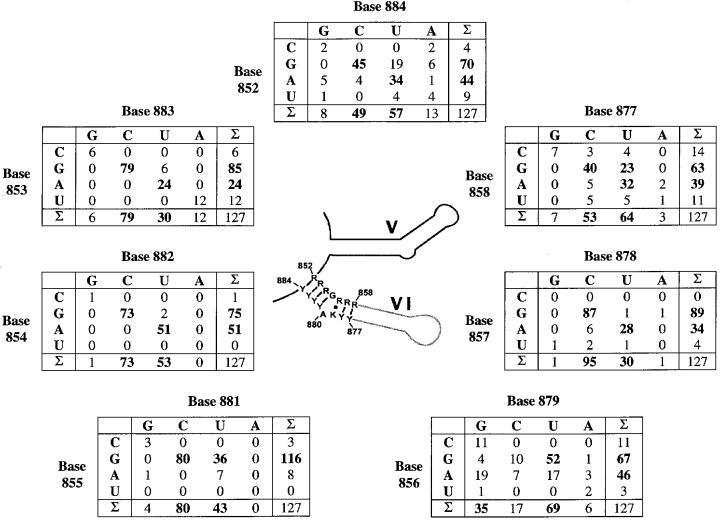
Fig. 7. Covariation of D6 nucleotides. The secondary structure consensus of D5 and D6 are shown in the center. The numbering of nucleotides is derived from the ai5γ nomenclature. Each table displays the co-variation of 2 nt putatively involved in base pairing around the branching adenosine. The D56 alignment from which these matrices were calculated contains 127 sequences belonging to fungal, plant and algae organelles, and bacteria. All were downloaded from DDBJ/EMBL/GenBank and aligned combining the comparative approach with thermodynamic methods (Mfold server, Zuker, 1995–2000, Rensselaer Polytechnic Institute). Introns were classified by gene family and parsed into groups IIA and IIB according to standardized parameters (Michel et al., 1989). This resulted in the inclusion of 68 group IIB introns, 45 IIA introns and 14 unclassified bacterial introns that adopt features from both subgroups. Seven introns belonging to group IIA were excluded due to the lack of a bulged adenosine. Five of these are inserted into chloroplastid tRNAVal genes (trnV, J.Vogel, personal communication) and two into chloroplastic protease genes [clpP/2 or ORF203/2, sequence numbers 24 and 25 in Michel et al. (1989)]. Alignment of all sequences is provided as Supplementary data available at The EMBO Journal Online.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.