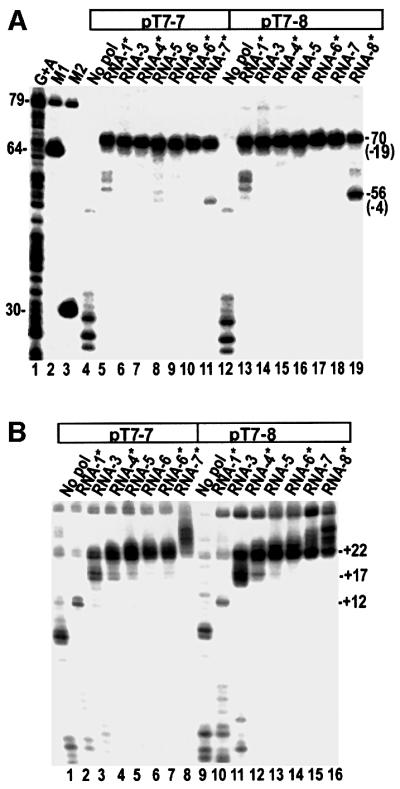
Fig. 1. (A) Exo III mapping of the upstream boundary of T7RNAP ITCs formed with either 5′-template strand-labeled pT7-7 (lanes 4–11) or pT7-8 (lanes 12–19). The length of the RNA that can be made, based on the NTPs present, is specified above each lane. Reactions in lanes 5 and 13 (‘RNA-1*’) contain 3′-dGTP only. In lanes marked with an asterisk, the RNA lacks a 3′-OH group and the incoming NTP is present in the reaction at 1 mM, so that complexes with bound NTP can form. Lane 1, Maxam–Gilbert (G+A) sequencing reaction with labeled pT7-7; lanes 2 and 3, restriction digests of pT7-7. DNA fragment lengths are as indicated, and the positions at which exo III digestion is halted (relative to the transcription start site) are in parentheses. (B) λ exo digestion of the downstream boundary of T7RNAP ITCs formed with either 3′-template strand-labeled pT7-7 (lanes 1–8) or pT7-8 (lanes 9–16).
