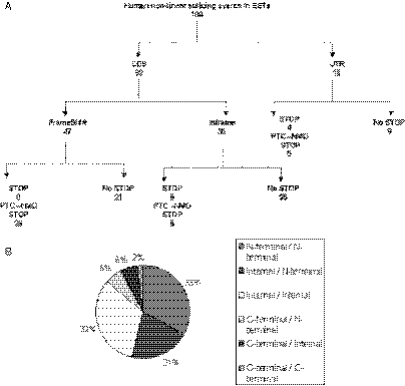
Figure 3.
An analysis of a representative sample of 100 human non-linear splicing events in EST sequences. (A) Open reading frame analysis of 100 human non-linear spliced EST sequences. CDS: non-linear splice site involves only exons within the coding sequence of the gene. UTR: non-linear splice site involves exons within the untranslated region of the gene. FrameShift: the non-linear splice introduces a frame shift in the open reading frame of the sequence when compared to the reference protein sequence for the gene. Inframe: the non-linear splice conserves the open reading frame of the sequence when compared to the reference protein sequence for the gene. STOP: the non-linear splice introduces a premature stop codon in the sequence. PTC-NMD STOP: the non-linear splice introduces a premature stop codon in the sequence that is >50 nt upstream of the final exon and is therefore a candidate sequence for nonsense-mediated RNA decay. (B) Summary of the non-linear splice locations in the proteins of 100 human events. The potential protein sequence regions affected by the 100 human non-linear human events in EST sequences. Each protein sequence was divided into thirds by the number of amino acids. The locations of the non-linear splice within the open reading frame of the protein were classified as N-terminal when occurring in the first third of the protein sequence, internal when occurring in the second third and C-terminal when occurring in the last third of the protein sequence.