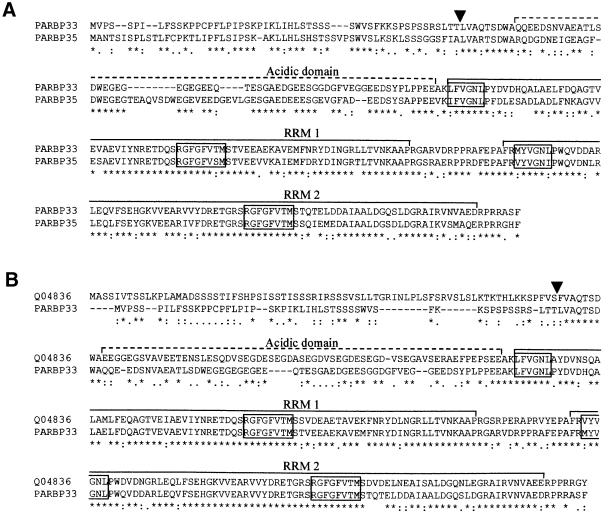
Fig. 6. Sequence alignments between the avocado chloroplast RNA-binding proteins PARBP33 and PARBP35 (A) and between PARBP33 and ATRBP33 from A.thaliana (Swiss-Prot database accession No. Q04836) (B). The alignments were obtained using the Clustal W program with opening gap and end gap penalties of 10, and extending gap and separation gap penalties of 0.05. Arrowheads point to the processing site that in ATRBP33 separates the transit peptide from the mature protein (Ohta et al., 1995) and the putative processing sites in PARBP33 and PARBP35. Dashed and continuous lines delimit the acidic domain and the two RNA recognition motifs (RRMs), respectively. Conserved RNP-1 octapeptide and RNP-2 hexapeptide in both RRMs are boxed. ‘*’ indicates identical residues in the alignment, and ‘:’ and ‘.’ indicate conserved and semi-conserved substitutions, respectively.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.