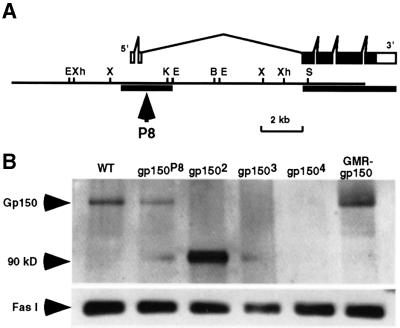
Fig. 1. Identification of loss-of-function alleles of the gp150 gene. (A) The gp150 gene consists of six exons and five introns with the complete coding sequence restricted within a 4 kb genomic region. The filled boxes represent a complete 3156 bp ORF. Sequences of two genomic regions indicated by the thick horizontal lines were determined to illustrate intron–exon structures of the gp150 transcriptional unit. A PCR method was used to obtain the downstream genomic sequence of gp150 that was not included in the genomic clone. In P8, a P transposon was inserted in the second intron of gp150. Restriction enzymes used for mapping include BamHI (B), EcoRI (E), KpnI (K), SalI (S), XbaI (X) and XhoI (Xh). (B) A western blot was probed with a Gp150 antibody that was made against the N-terminal region (amino acids 5–192) of Gp150. Since no Gp150 protein was detectable in gp1502, gp1503 and gp1504 mutants, these alleles can be considered as molecular nulls. The gp1502 mutation results in the production of a non-functional 90 kDa protein. This is based on observations that gp1502 is recessive and the mutant phenotypes caused by gp1502 are similar to that of gp1503 and gp1504. The 70 kDa Fas I protein served as an internal control and was detected by anti-Fas I (MAb6D8) antibody in all lanes.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.