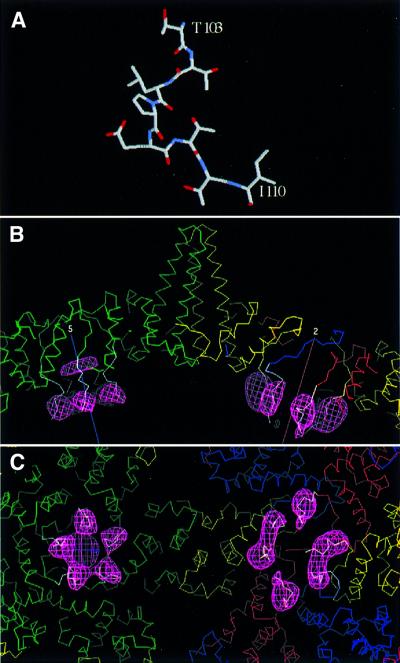
Fig. 6. Modeling a peptide structure into linker-associated densities visualized by cryo-EM difference imaging. Residues 103–110 of cellobiose dehydrogenase (Hallberg et al., 2000) are almost completely identical to the linker sequences (A). In (B), this peptide is shown (white α-carbon trace) docked into the difference density (pink mesh) from the T = 4 capsids. Quasi-equivalent core domain subunits are shown in green, yellow, red and blue. Five-fold (at left) and 2-fold (at right) symmetry axes are marked. In (C), the view is from the inside of the capsid. We propose that a semi-flexible hinge allows the linker to pivot, so that it serves as a spacer that prevents the protamine domains from intruding too closely into inter-core domain contacts, and also confers mobility on the protamine domains’ interactions with nucleic acid.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.