Abstract
The CCR4–NOT complex from Saccharomyces cerevis iae is a general transcriptional regulatory complex. The proteins of this complex are involved in several aspects of mRNA metabolism, including transcription initiation and elongation and mRNA degradation. The evolutionarily conserved CCR4 protein, which is part of the cytoplasmic deadenylase, contains a C-terminal domain that displays homology to an Mg2+-dependent DNase/phosphatase family of proteins. We have analyzed the putative enzymatic properties of CCR4 and have found that it contains both RNA and single-stranded DNA 3′–5′ exonuclease activities. CCR4 displays a preference for RNA and for 3′ poly(A) substrates, implicating it as the catalytic component of the cytoplasmic deadenylase. Mutations in the key, conserved catalytic residues in the CCR4 exonuclease domain abolished both its in vitro activities and its in vivo functions. Importantly, CCR4 was active as a monomer and remained active in the absence of CAF1, which links CCR4 to the remainder of the CCR4–NOT complex components. These results establish that CCR4 and most probably other members of a widely distributed CCR4-like family of proteins constitute a novel class of RNA–DNA exonucleases. The various regulatory effects of the CCR4–NOT complex on gene expression may be executed in part through these CCR4 exonuclease activities.
Keywords: CCR4–NOT complex/deadenylase/exonuclease/mRNA degradation/transcription
Introduction
The CCR4 protein from Saccharomyces cerevisiae is an evolutionarily conserved protein, and has orthologs in all higher eukaryotic organisms examined to date. In yeast, the CCR4 protein has been identified in the CCR4–NOT protein complex, which exists in two forms in vivo: 1.0 and 1.9 MDa. The smaller core complex, which has been purified (Liu et al., 1998; Bai et al., 1999; Chen et al., 2001), contains the following proteins: CCR4, CAF1, five NOT proteins (NOT1–NOT5), CAF40 and CAF130.
The CCR4 gene was identified initially by mutations that suppressed spt10-enhanced ADH2 expression and affected the derepression of ADH2 and other non-fermentative genes (Denis, 1984). However, it has become clear since then that CCR4, CAF1 and the NOT proteins affect diverse processes in yeast and do so both positively and negatively (Denis and Malvar, 1990; Sakai et al., 1992; Collart and Struhl, 1993, 1994; Liu et al., 1998, 2001; Bai et al., 1999). CCR4 and its associated proteins have been implicated in multiple roles in the control of mRNA metabolism, including initiation, elongation and mRNA degradation (Badarinarayana et al., 2000; Lemaire and Collart, 2000; Daugeron et al., 2001; Denis et al., 2001; Tucker et al., 2001).
Biochemical studies in our laboratory have shown that the CCR4 protein contains three major functional domains (Draper et al., 1994). The N-terminal region, which is rich in glutamines and asparagines, contains an activation domain that may interact with the transcriptional machinery. The central region of the protein (residues 350–473) contains five tandem copies of a leucine-rich repeat (LRR) domain (Malvar et al., 1992). This domain has been shown to interact with CAF1 (Draper et al., 1995) and other putative components of the 1.9 MDa CCR4–NOT complex (Liu et al., 1997, 2001) and to be the region of CCR4 that links CCR4 to the 1.0 MDa CCR4–NOT complex (Draper et al., 1994; Liu et al., 1998). Bioinformatic studies that we and others have carried out have shown that the C-terminal region of the CCR4 protein displays homology to apurinic (AP) endonucleases. AP endonucleases are a group of critical enzymes that maintain genome integrity against the spontaneous production of abasic (AP) sites. Three representative AP endonucleases in this homologous family are exonuclease III (ExoIII) from Escherichia coli, APE1 (HAP1) from human and APN2 from the yeast S.cerevisiae. This family of Mg2+-dependent endonucleases is also homologous to inositol polyphosphate-5′-phosphatases and sphingomyelinases (Dlakic, 2000; Whisstock et al., 2000).
The CAF1 protein, which is tightly associated with CCR4, is also required for glucose derepression of gene expression (Sakai et al., 1992; Draper et al., 1995; Hata et al., 1998). The yeast CAF1 protein orthologs in Mus musculus and Caenorhabditis elegans (Draper et al., 1995), but not yeast CAF1 itself, have been suggested to contain a putative exonuclease domain (3′–5′ exoribonuclease or/and 3′–5′ exodeoxyribonuclease) by hidden Markov model and phylogenetic studies (Moser et al., 1997). This evidence and a recent publication (Daugeron et al., 2001) postulate that yeast CAF1 might possess an extremely divergent exonuclease domain that has a tertiary structure and/or activity similar to that of metazoan CAF1 (Moser et al., 1997). However, mutations in the key catalytically conserved residues of yeast CAF1 did not impair its physiological function in vivo (our unpublished observations).
The first direct physical evidence of an enzymatic function for the CCR4–NOT complex was the demonstration that CCR4 and CAF1 are components of the major cytoplasmic mRNA deadenylase in S.cerevisiae (Daugeron et al., 2001; Tucker et al., 2001). ccr4 and caf1 strains showed defects in both the rate and extent of deadenylation for the MFA2pG and PGK1pG mRNAs. In addition, a ccr4 deletion together with a pan2 (another known deadenylase) deletion completely blocked the deadenylation process, indicating that these two genes are responsible for most if not all of the deadenylase activity in yeast. Further, FLAG-CAF1 co-purified with a poly(A)-specific deadenylase activity, and this activity required the presence of the CCR4 protein (Tucker et al., 2001).
In the present research, we present direct biochemical evidence that indicates that CCR4 is a 3′–5′ RNase and single-stranded (ss)DNase with a marked preference for poly(A) substrates. Mutations in the putative key, catalytically important exonuclease residues of CCR4 abrogate CCR4 enzymatic activity and, correspondingly, CCR4 functions in vivo. Moreover, CCR4 was active as a monomer and remained active in the absence of CAF1 and therefore the rest of the CCR4–NOT complex components. These findings strongly support the designation of the CCR4 protein as the catalytic subunit of the cytoplasmic mRNA deadenylase and define the CCR4-like family of proteins as a novel class of exoribonuclease.
Results
The CCR4 protein is a member of the Mg2+-dependent ExoIII/HAP1 (APE1) nuclease family
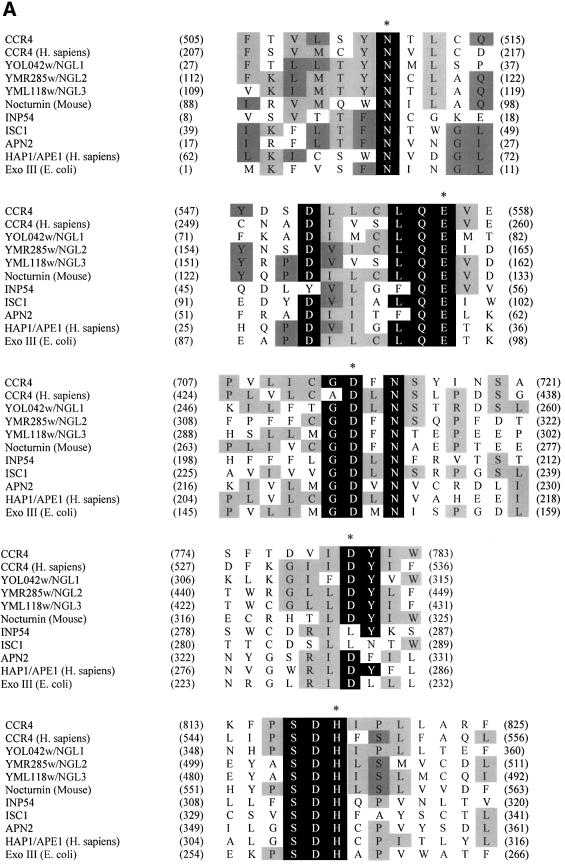
The C-terminal region of CCR4 has been shown to share homology with a number of eukaryotic proteins such as angel and nocturnin (Green et al., 1996; Kurzik-Dumke and Zengerle, 1996; Dupressoir et al., 1999). Deletion of this region in CCR4 inactivates the protein (Draper et al., 1994). To search for potential functional domains within the C-terminal region of CCR4, we performed PSI-BLAST searches of the non-redundant protein database using the amino acid sequence of the CCR4 C-terminal domain (495–837) as a probe. The C-terminal domain of the CCR4 protein shared high homology with ExoIII (E.coli) and HAP1 (APE1) (Homo sapiens), an AP endonuclease (Figure 1A). Independently, Dlakic (2000) and Whisstock et al. (2000) have shown this family of proteins to include sphingomyelinases and inositol polyphosphate-5′-phosphatases. In S.cerevisiae, there exists a group of CCR4 C-terminal-like proteins, namely YOL042w (NGL1), YMR285w (NGL2) and YML118w (NGL3), as well as an AP endonuclease (APN2) and putative sphingomyelinases and inositol polyphosphate phosphatases.
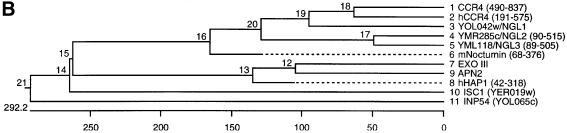
Fig. 1. Sequence alignment of CCR4 protein within the Mg2+-dependent ExoIII/APE1 (HAP1) nuclease family. (A) By performing Clustal W 1.8and PIMA multiple sequence alignment together with Block Maker search, we aligned the C-terminal domain of CCR4 protein, its yeast homologs (YOL042w, YMR285w and YML118w) and human ortholog (KIAA1194) with ExoIII, HAP1 (APE1), APN2 (yeast ortholog of AP endonuclease), ISC1 (yeast sphingomyelinase) and INP52 (one of the representative yeast inositol 5′-phosphatases). The most conserved catalytic amino acid residues are highlighted with a black background. The critical catalytic amino acids for enzymatic reactions are also marked with an asterisk at the top of each motif. The residues with at least four amino acids conserved in the same position are highlighted with a gray background. (The amino acids leucine and isoleucine were considered to be identical for this analysis.) (B) Phylogenetic tree for the C-terminal domain of CCR4 protein and enzymatic domains from this nuclease family aligned by Clustal methods from DNA Star. In this phylogenetic tree, the numbers in the branch length represent approximately the divergence from an ancestral node (DNA Star Program Manual).
All of the above proteins share five highly conserved motifs as follows: I, SYNTL; II, CLQE; III, GDFN; IV, IDYI; and V, PSDH (Figure 1A). Based on the high resolution X-ray crystal structure of ExoIII (Mol et al., 1995) and the human APE1 in complex with abasic DNA (Gorman et al., 1997; Mol et al., 2000), all five motifs play critical roles for AP endonuclease activity. For APE1, the hisidine residue (marked with an asterisk in Figure 1A) in motif V makes a direct hydrogen bond with the target AP site 5′-phosphate and in turn is stabilized by hydrogen bonding to an aspartate residue (an asterisk in Figure 1A) in motif IV. Phosphodiester bond cleavage is achieved by hydroxyl nucleophile attack by the aspartate residue in motif III. This aspartate residue is oriented correctly by hydrogen bonds from the asparagine residue in motif I. The metal ion-binding motif II utilizes a glutamic acid residue to bind an Mg2+ ion, stabilize the transition state and facilitate the OH– leaving group (Wilson et al., 1997; Mol et al., 2000).
We generated a phylogenetic tree relating the C-terminal CCR4 region to other proteins containing these five motifs (Figure 1B). As expected, yeast CCR4 and human CCR4 were extremely closely related. The CCR4-like proteins in yeast (NGL1, NGL2 and NGL3) and mouse nocturnin share a high degree of sequence homology, whereas the nucleases ExoIII, APN2 and human HAP1 (APE1) are more distantly related. Sphingomyelinases and inositol polyphosphate-5′-phosphatases, such as ISC1 (yeast sphingomyelinase) and INP54 (one of the yeast inositol phosphatases), are more distantly related to the CCR4-like proteins and the nucleases. This analysis suggests that CCR4 is most likely to contain nuclease activities.
In addition to its AP endonuclease activities, ExoIII also displays related DNA/RNA enzymatic activities, which include 3′-phosphodiesterase and 3′-phosphomonoesterase activities, RNase H activity and 3′–5′ exonuclease activity. Similarly, HAP1 (APE1) protein, the major AP endonuclease in human cells, also displays the same variety of enzymatic activities related to DNA/RNA metabolism (Barzilay et al., 1995; Gorman et al., 1997; Lucas et al., 1999; Nguyen et al., 2000). We subsequently investigated the enzymatic functions of the CCR4 protein as suggested from this bioinformatic analysis.
Mutation of conserved amino acids in CCR4 impairs its in vivo function
In order to assess the importance of the nuclease domain to CCR4 function, we mutated four of the most conserved putative catalytic residues in the CCR4 protein that are located in motifs II–V of this nuclease/phosphatase family. These point mutations and their respective alleles are designated ccr4-1 (E556A), ccr4-2 (D713A), ccr4-3 (D780A) and ccr4-4 (H818A).
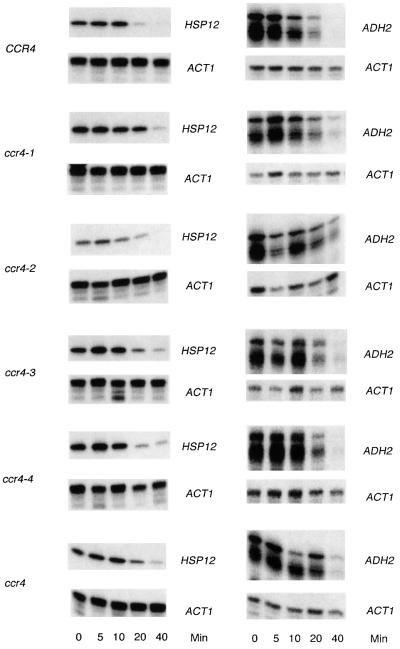
The mutated genes were integrated respectively at the TRP1 locus in a strain containing a ccr4-10 allele that does not produce a CCR4 protein in vivo (Draper et al., 1994). All four mutated proteins were expressed at levels comparable to the wild-type CCR4 (data not shown). Moreover, as expected, all of the four mutated proteins interacted quite well with CAF1, as assayed by both two-hybrid and immunoprecipitation analysis (Table I), confirming their intact expression in yeast and consistent with the model that it is the LRR of CCR4 that mediates contact with CAF1 (Draper et al., 1995). Mutation of the key Mg2+-binding residue (E556A), which blocks endonuclease activity in other members of this family, abrogated CCR4 function in vivo, resulting in caffeine sensitivity, inability to grow at low temperature and a defect in non-fermentative growth at 37°C (Table I). Similarly, mutation of the histidine residue (H818A), which is involved in stabilizing the phosphate during catalysis, and the other catalytically important residues (D780A and D713A) significantly reduced CCR4 function (Table I). We subsequently assessed the effect of these mutations on the rate of degradation of three mRNAs: ADH2, HSP12 and GAL1 mRNA (Table II; Figure 2). In general, the ccr4-1, -2, -3 and -4 alleles reduced the rate of degradation of ADH2, HSP12 and GAL1 by 1.4- to 1.6-fold, comparable with the 1.4- to 1.7-fold effects of deleting ccr4 on their mRNA half-lives (Table II). Using a set of similar catalytically inactive mutations, it has been shown recently that such mutations also fail to deadenylate mRNA fully in vivo (Tucker et al., 2002). These results indicate that the residues of the CCR4 C-terminal domain that are functionally homologous to those present in the ExoIII nuclease family are critically important to CCR4 functions in vivo.
Table I. Mutations in the conserved catalytic residues severely affect physiological functions of CCR4 in vivo.
| Binding toCAF1 | Single copy of CCR4 |
High copy of CCR4 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| YD(30°C) | Caffeine(8 mM) | Glycerol(37°C) | YD(13°C) | YD(30°C) | Caffeine(8 mM) | Glycerol(37°C) | YD(13°C) | ||
| CCR4 | ++ | + | + | + | + | + | + | + | + |
| ccr4-1 (E556A) | ++ | + | – | – | w | + | – | – | – |
| ccr4-2 (D713A) | ++ | + | – | – | – | + | – | – | – |
| ccr4-3 (D780A) | ++ | + | – | – | w | + | + | w | – |
| ccr4-4 (H808A) | ++ | + | – | – | w | + | – | – | – |
| ccr4 | – | + | – | – | – | + | – | – | – |
Single copy of CCR4 refers to the following strains: ccr4 strain (MD9-7c); CCR4 strain (MD9-7c-120); ccr4-1 strain (MD9-7c-120-1); ccr4-2 strain (MD9-7c-120-2); ccr4-3 strain (MD9-7c-120-3); and ccr4-4 strain (MD9-7c-120-4). High copy of CCR4 represents LexA–CCR4 and its derivative mutants that were expressed in MD9-7c, which carries the ccr4-10 allele that does not produce a CCR4 protein in vivo (Draper et al., 1994). Growth was scored on the indicated growth plates as: + = good; w = weak, – = none. Binding to CAF1 was monitored by using the ability of LexA–CCR4 fusions to bind B42-CAF1 (Draper et al., 1995) and confirmed by the ability of the CCR4 variants to immunoprecipitate CAF1 in the MD9-7c series of strains.
Table II. Effect of ccr4 alleles on mRNA half-lives.
| Alleles | mRNA half-lives (min) |
||
|---|---|---|---|
| ADH2 | HSPn12 | GAL1 | |
| CCR4 | 11 | 9 | 8.0 |
| ccr4-1 (E556A) | 16 | 12 | 11 |
| ccr4-2 (D713A) | 17 | 20 | 13 |
| ccr4-3 (D780A) | 15 | 13 | 10 |
| ccr4-4 (H808A) | 11 | 13 | 13 |
| ccr4 | 19 | 13 | 12 |
Strains are the same as described in Table I. mRNA half-lives were determined in the single-copy MD9-7c strains by quantitative S1 nuclease protection assays. RNA was extracted for times up to 40 min from yeast pre-grown for 3 h on YEP medium containing 2% glycerol/20% ethanol for ADH2 and HSP12, and 2% galactose/2% raffinose for GAL1 that was shifted to fresh medium containing 8% glucose. For the GAL1 determination, the standard errors of the mean were <5% for CCR4, 10% for ccr4-3 and 15–20% for the other alleles. For ADH2 and HSP12, the standard errors of the mean were <10% for all alleles, with the exception of <20% for ccr4-2 (ADH2) and ccr4-3 (ADH2), and <30% for ccr4-2 (HSP12).
Fig. 2. mRNA decay of HSP12 and ADH2. RNA was isolated at the time point indicated following shifting of yeast from medium containing ethanol/glycerol to repressive medium containing glucose. HSP12 and ADH2 transcription are shut off upon growth on glucose-containing medium. Quantitative S1 nuclease protection assays were conducted to measure mRNA levels. ACT1 mRNA is used to standardize loading as its synthesis is unaffected by growth on glucose-containing medium. Decay rates for HSP12 were calculated without including the zero time point since HSP12 synthesis does not cease immediately.
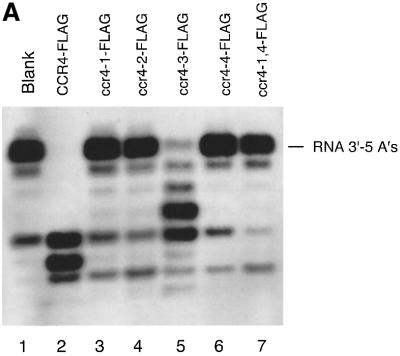
When each of the mutated CCR4 proteins was overexpressed in the cell as LexA fusion proteins, we found, however, that LexA–ccr4-3 (D780A) was able partially to complement a ccr4 deletion (Table I). This result suggests that the ccr4-3 protein retains a significant level of catalytic activity, as confirmed below (see Figures 4B and 5A). This result is consistent with the model that the D780 residue plays an indirect role in catalysis (presumably stabilizing H818), while the other three residues, E556, D713 and H818, are predicted to be directly involved in catalysis (Mol et al., 2000).
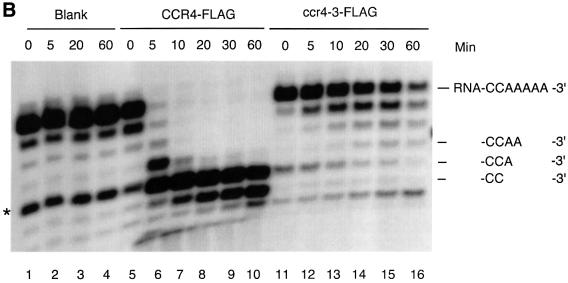
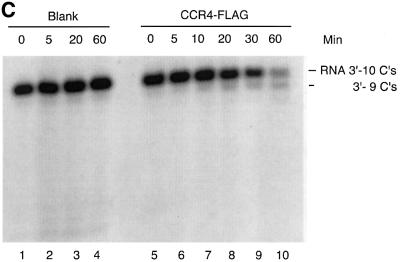
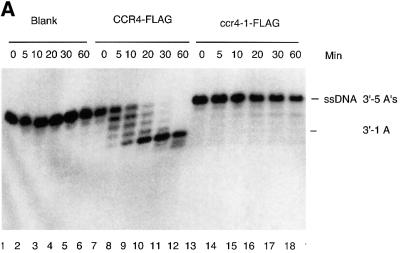
Fig. 4. The CCR4 protein is a poly(A) exoribonuclease. (A) Time course of CCR4-FLAG fusion protein degradation of 5′-labeled RNA with 20 As at its 3′ end as substrate. Radioactively labeled RNA substrate was analyzed on a denaturing sequencing gel. The three minor RNA species migrating below the full-length RNA oligonucleotides (see lane 1) most probably represent oligonucleotides lacking one, two and three As, respectively, as each of these substrates was degraded completely by CCR4-FLAG (lanes 8–10). CCR4-FLAG and ccr4-1-FLAG protein abundance was 32 ng in each reaction for lanes 6–15. CCR4-FLAG and the variant proteins were extracted from KY803-1a (ccr4) strains. The indicated times refer to minutes of incubation prior to stoppage of the enzyme reaction. The dosage effect of CCR4-FLAG enzymatic activities was determined following 20 min of incubation. Lanes 1–5, no protein extract was added to RNA substrate; lanes 6–10, CCR4-FLAG extracts; lanes 11–15, ccr4-1-FLAG extracts; lanes 16–18, CCR4-FLAG extracts containing no CCR4-FLAG, or 16 or 32 ng of CCR4-FLAG; lanes 19 and 21, ccr4-1-FLAG extracts at the same respective concentrations as indicated for lanes 16–18. (B) Time course of CCR4-FLAG and ccr4-3-FLAG fusion protein degradation of 5′-labeled RNA with five As at its 3′ end. Reactions were conducted as described in (A). An additional RNA species that migrated four nucleotides smaller than the full-length substrate (see lane 1) is discussed in the text. (C) Time course of CCR4-FLAG fusion protein enzyme activity with 5′-labeled RNA with 10 Cs at its 3′ end.
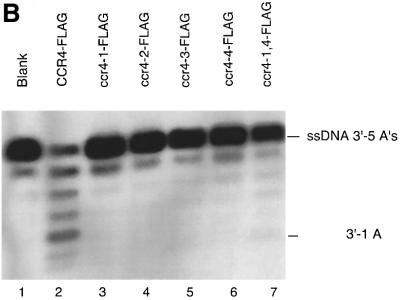
Fig. 5. The effect of CCR4 mutations on its enzymatic activity. (A) ccr4 mutations block CCR4 poly(A) RNA exonuclease activity. Reaction conditions were as described in Figure 4, except that the incubation time was 60 min. The CCR4 protein variants are described in the text, and ccr4-1,4-FLAG contains both D556A and H818A alterations. The RNA substrate was the 27mer oligonucleotide with five As at its 3′ end. (B) CCR4-FLAG mutants block CCR4 exonuclease activity. The substrate was the RNA containing 10 Cs at its 3′ end. The reaction conditions were the same as in Figure 3A.
We also noted that this C-terminal region defines a unique and independent domain of CCR4. When this region was overexpressed by itself as a LexA fusion, it could partially suppress the ccr4 37°C glycerol growth defect; LexA alone does not complement CCR4 function (Draper et al., 1994; data not shown). Importantly, this ability of the C-terminal domain to complement the glycerol growth defect was compromised when the ccr4-1 (E556A) variant was analyzed.
The CCR4 protein displays 3′–5′ poly(A) exoribonuclease activity
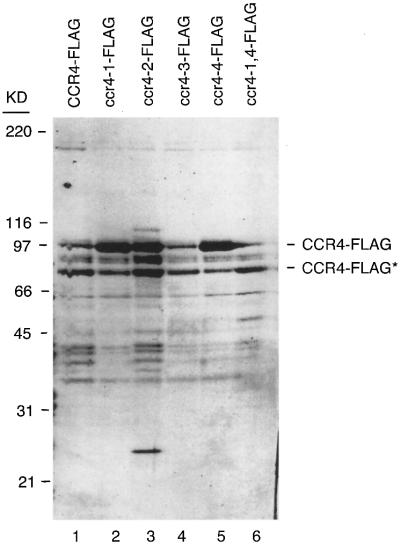
In order to examine CCR4 enzymatic activities in vitro, we constructed full-length CCR4 protein and each of the above-described mutated CCR4 protein derivatives with a FLAG tag at their C-terminal ends. Upon transformation into a ccr4 deletion strain, the CCR4-FLAG derivatives complemented ccr4 in the same manner as the respective CCR4 derivatives (data not shown; Table I). We purified the CCR4-FLAG fusion proteins following a one-step affinity chromatography. Coomassie Blue staining of our purified material indicated that CCR4-FLAG represented 20–40% of total extracts (Figure 3). Gel filtration chromatography analysis of crude extracts indicated that while most of the CCR4-FLAG protein was monomeric, a significant amount of CCR4-FLAG was in its normal 1.9 and 1.0 MDa complexes (data not shown). Correspondingly, western analysis also showed that our purified material, whether derived from CCR4-FLAG- or ccr4 mutant-FLAG-containing strains, contained comparable amounts of the other CCR4–NOT components, such as CAF1, CAF40 and the NOT1–NOT5 proteins (data not shown). The CCR4-FLAG protein derivatives were assayed subsequently for their enzymatic activities on several DNA/RNA substrates.
Fig. 3. Coomassie Blue-stained SDS–polyacrylamide gel of CCR4-FLAG preparations. The band marked with the asterisks (CCR4-FLAG*) lacks ∼100 amino acids of the extreme N-terminus of CCR4 as determined by western analysis. Loss of this region of CCR4 had no effect on CCR4 in vivo activity (Draper et al., 1994). Molecular weight standards are indicated on the left and the respective CCR4-FLAG mutants are indicated.
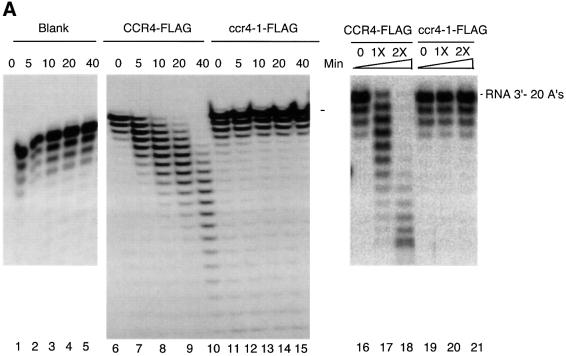
Previously, the CCR4–NOT complex, isolated using a tagged CAF1 fusion, was shown to contain a mRNA deadenylase activity (Tucker et al., 2001). The component displaying this activity was not, however, identified. We directly assayed CCR4-FLAG for poly(A) deadenylase activity by incubating purified CCR4-FLAG with a 30mer RNA oligonucleotide with 20 As at its 3′ end. After incubation of this poly(A) RNA substrate with CCR4-FLAG for 5 min, we identified a ladder pattern of deadenylated RNA species on the denaturing sequencing gel (Figure 4A, compare lane 7 with 6). With increased incubation time, CCR4-FLAG resulted in increased abundance of the shorter species (lanes 8–10). Since this substrate was 5′-end-labeled, these smaller products represent shorter RNA species that resulted from trimming of the 3′ end. As shown in Figure 4A, up to 19 As were able to be removed from the 3′ end of the substrate. Control experiments indicated that the enzyme retained its activity for at least 1 h and that the enzyme activity was proportional to the concentration of CCR4-FLAG that was present (Figure 4A, lanes 16–18). In addition, using extracts without the CCR4-FLAG plasmid, we did not observe any enzymatic function (data not shown). In contrast to CCR4-FLAG, the ccr4-1 mutant protein, containing the E556A alteration which should abrogate CCR4’s ability to bind the Mg2+ ion, did not degrade this RNA substrate (Figure 4A, lanes 11–15). This analysis indicated that CCR4-FLAG is an exoribonuclease and can work on substrates containing at least 20 As.
We repeated this analysis employing a 27mer RNA oligonucleotide substrate with only five As at its 3′ end in order to observe whether CCR4 continued to degrade the RNA when it reached sequences containing residues other than an ‘A’. Within 5 min, CCR4-FLAG converted this substrate to an oligonucleotide containing only two As or one A at its 3′ end (Figure 4B, lane 6). Following longer incubation times, the terminal A was slowly removed (lanes 7–10). The slowed rate of removal of this terminal A was not affected by the length of incubation, since, as indicated above, the CCR4-FLAG protein did not display noticeable loss of enzymatic activity for at least 1 h under the conditions analyzed (Figure 4A; data not shown). These results demonstrate that the CCR4-FLAG protein displays 3′–5′ poly(A) exoribonuclease activity with preference towards substrates containing at least two As at its 3′ end.
Careful analysis of the gel in Figure 4B, lane 10, indicates that an additional band is formed below the RNA species marked ‘-CC-3′’. While this band could result from the 3′-C being removed by CCR4-FLAG, it could be derived from the non-full-length band in lane 1 that is marked with an asterisk. Relatedly, the formation of this band below ‘-CC-3′’ occurred with all ccr4 mutant derivatives (Figures 4B and 5A), indicating that the formation of this band occurred in a CCR4-independent manner, apparently from a contaminating nuclease. In order to investigate clearly whether CCR4 is able to degrade RNA with a defined C at its 3′ end, we used an RNA substrate with the same leading sequence as the oligonucleotide containing 20 As at its 3′ end, but which contained 10 Cs at its 3′ end. After a 60 min incubation, we could identify two, possibly three nucleotides being removed from the input substrate (Figure 4C, lanes 9 and 10; also Figure 5B, lane 2). Other results indicated that CCR4 was, however, incapable of degrading RNA substrates with a defined U or G at their 3′ end (data not shown). Based on these observations, the CCR4 protein is a 3′ exoribonuclease that displays a marked preference for poly(A) RNA substrates. Other experiments also indicate that capping of the mRNA does not affect CCR4 activity (Tucker et al., 2002).
Mutations in the conserved catalytic residues of CCR4 abrogate CCR4 enzymatic function
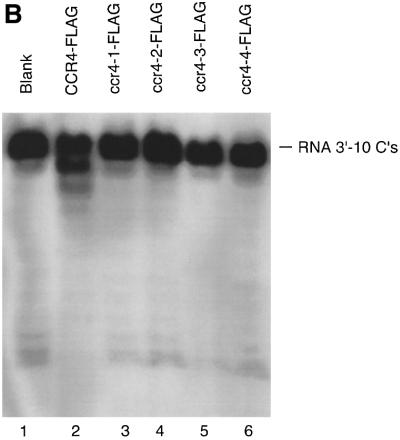
To assess the in vivo significance of this 3′–5′ exonuclease activity, we assayed the four ccr4 mutants, as well as one double mutant (ccr4-1,4), for their exoribonuclease activities. As shown in Figure 5A, using the RNA oligonucleotide substrate with five As, CCR4-FLAG and ccr4-3 (D780A)-FLAG were the only proteins that could degrade the RNA substrate, although the ccr4-3 (D780A) protein was much less active than wild-type CCR4. These results correlate directly with the in vivo phenotypes displayed by each of these mutants (Table I). Importantly, the ccr4-3 protein displayed some activity and when expressed at high levels in yeast could partially complement ccr4 function. We subsequently compared CCR4-FLAG and ccr4-3-FLAG enzyme activities directly as a function of time. As shown in Figure 4B, ccr4-3 did function as an exonuclease, albeit at a rate ∼50-fold less than CCR4, as determined by densitometric analysis of the data presented in Figure 4B. None of the ccr4 mutated proteins was functional with the RNA substrate containing 10 Cs at its 3′ end (Figure 5B).
CCR4 protein is also a 3′–5′ poly(dA) ssDNA exonuclease
Because the other nucleases in the APE1 family are active with DNA, CCR4-FLAG was also assayed for its enzymatic activity with DNA substrates. For an ssDNA substrate containing the same sequence as the RNA oligonucleotide with five As at its 3′ end, we found that CCR4-FLAG was able to shorten the input ssDNA to produce a product that was four nucleotides shorter (Figure 6A, lanes 7–12). This pattern of degradation was similar to that observed for the comparable RNA substrate (Figure 4B), except that the rate of the reaction was found to be 10- to 20-fold slower for the ssDNA substrate than for the RNA substrate. Moreover, we observed the accumulation of substrates lacking four nucleotides, which indicated that the CCR4-FLAG preferred the ssDNA substrate with at least two A residues at its 3′ end. It can also be observed in Figure 6A that CCR4-FLAG can remove this last A very slowly. In contrast, the ccr4-1 mutant protein was no longer capable of degrading this ssDNA substrate (Figure 6A, lanes 13–18). The other above-described CCR4-FLAG mutant proteins were also incapable of degrading this ssDNA substrate (Figure 6B). In addition, this enzymatic function was totally dependent on CCR4-FLAG protein. In the extracts without CCR4-FLAG plasmid, we did not observe any enzymatic activity (data not shown).
Fig. 6. CCR4 is an ssDNA exonuclease. (A) Time course of CCR4-FLAG enzyme activity with the ssDNA substrate containing five As at its 3′ end. A 160 ng aliquot of CCR4-FLAG and of ccr4-1-FLAG was used in the incubations. (B) ccr4 mutations block CCR4 ssDNA exonuclease activity. A 32 ng aliquot of each CCR4-FLAG variant was used and the reactions were for 60 min.
We also analyzed an ssDNA substrate with a 3′-CG end. We did not observe, however, any degradation of the input ssDNA (data not shown). Importantly, we did not observe any enzymatic activity using double-stranded (ds)DNA substrates, whether it be exo- or endonucleolytic cleavage (data not shown). CCR4 was also unable to cleave RNA, ssDNA or dsDNA substrates internally when they contained 10 consecutive As located within the polynucleotide sequences (data not shown).
CAF1 is not required for CCR4-FLAG exoribonuclease activity
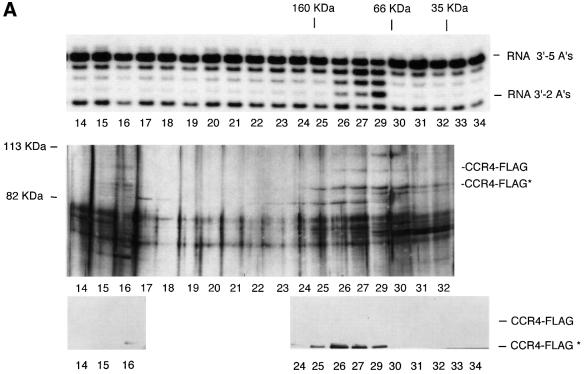
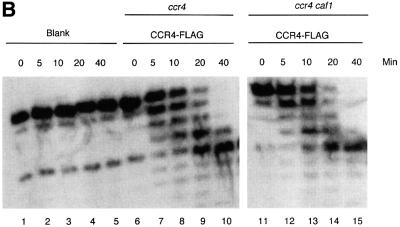
The above results indicated that CCR4-FLAG contains RNA and ssDNA exonuclease activity and that mutations in the catalytically important residues of the CCR4 exonuclease domain block this activity. It remains formally possible, however, that CAF1, a 3′–5′ exonuclease (Moser et al., 1997; Daugeron et al., 2001), could be the enzyme responsible for the activity and that the CCR4 mutations somehow affect CAF1 activity. We tested whether CCR4 was sufficient for the RNase activity by two approaches. First, we subjected the CCR4-FLAG material to an additional chromatographic step. Separation of the CCR4-FLAG material on a Superdex 200 gel filtration column showed that CCR4-FLAG, migrating at 97 and 84 kDa, co-eluted with the RNase activity (Figure 7A, fractions 26–29). Fraction 28 material was lost in this particular experiment, but other experiments confirmed that the enzymatic activity migrated in fractions 27–29, with a peak of activity at fraction 28. CCR4-FLAG was, therefore, enzymatically active in the absence of the CCR4–NOT complex. Secondly, we analyzed CCR4- FLAG activity from a strain containing a caf1 deletion. CCR4-FLAG isolated from a caf1 deletion strain was found to be just as active in degrading the RNA substrate containing five As at its 3′ end as CCR4-FLAG from a CAF1 strain. (Figure 7B). Since CAF1 is absolutely required for CCR4 association with the NOT1–NOT5, CAF40 and CAF130 proteins (Liu et al., 1998; Bai et al., 1999; Chen et al., 2001), our results indicate that the other components of the CCR4–NOT complex are not required for CCR4-FLAG enzymatic activity in vitro. These results confirm that CCR4 is the enzymatically active component of the CCR4-CAF1 deadenylase and agree with the previous observation that CCR4 is required for the deadenylase activity observed in a FLAG-CAF1 preparation (Tucker et al., 2001).
Fig. 7. CAF1 protein is not required for CCR4 nuclease activity. (A) Deadenylase activity co-migrates with CCR4-FLAG. Purified CCR4-FLAG was subjected to Superdex 200 gel filtration chromatography. Top panel: determination of deadenylation activity using the 27mer RNA substrate with five As at its 3′ end. Middle panel: silver staining profile of proteins. CCR4-FLAG and CCR4-FLAG* are indicated. Bottom panel: western analysis using FLAG antibody. In the original western blot, CCR4-FLAG could be detected in fractions 26 and 27. Molecular weight standards at the top of the figure refer to IgG (160 kDa), BSA (66 kDa) and lactoglobulin (35 kDa). (B) CCR4-FLAG enzymatic activity in ccr4 and ccr4 caf1 strains. CCR4-FLAG plasmid was expressed in strains EGY188-1a (ccr4) (lanes 6–10) and EGY188-1a-1-c1 (ccr4 caf1) (lanes 11–15). Reaction conditions were as described in Figure 4.
Human CCR4-FLAG also displays 3′ exoribonuclease activity
The identification of mammalian orthologs to CAF1, CAF40 and NOT proteins, and the retention of the physical interaction between these human proteins, has suggested the existence of a human CCR4–NOT complex (Draper et al., 1995; Albert et al., 2000; Chen et al., 2001). In light of the above data indicating that CCR4 can function as a deadenylase, it is likely that the human CCR4 protein can also function as a deadenylase. Consequently, using a cDNA encoding one of the two human CCR4 genes (KIAA1944), we expressed human CCR4 in a yeast strain lacking the endogenous ccr4 gene. The human CCR4 protein, although expressed in yeast, was found to be unable to complement any of the phenotypes mentioned in Table I that typically are associated with a ccr4 deletion (data not shown). The reason for this functional inactivity might result from the inability of human CCR4 to associate physically with yeast CAF1, as determined by two-hybrid and immunoprecipitation studies (data not shown), or from a low level of deadenylase activity (see below). Human CCR4 did interact with mouse or human CAF1 that was co-expressed in yeast, suggesting that human CCR4 protein folded into a functionally intact protein in yeast (data not shown).
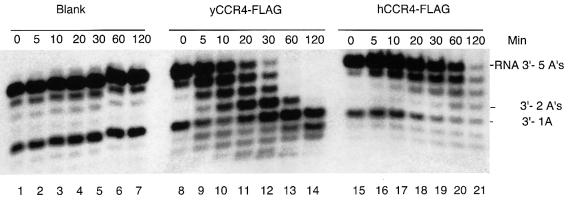
The enzymatic activity of human CCR4-FLAG was examined using the 27mer RNA substrate with five As described above. The yield of human CCR4-FLAG was much less than that found for yeast CCR4-FLAG, and Coomassie Blue staining of human CCR4-FLAG preparations indicated that human CCR4-FLAG constituted only 18% of the total protein extract (data not shown). Human CCR4-FLAG was capable of removing nucleotides from the RNA substrate over the course of the enzyme reaction (Figure 8, lanes 15–21), although it was much less active than yeast CCR4. These results confirm that human CCR4 is also an exoribonuclease, although improved preparations of the protein will be required to analyze its exact substrate preferences.
Fig. 8. Human CCR4-FLAG is an RNA exonuclease. Reaction conditions are as described in Figure 2. The indicated times refer to minutes of incubation prior to stoppage of the enzyme reaction. Lanes 1–7, no protein added; lanes 8–14, yeast CCR4-FLAG at 10 ng in each reaction; lanes 15–21, human CCR4-FLAG at 10 ng in each reaction. The RNA substrate contained five As at its 3′ end.
Discussion
The CCR4 protein is a 3′–5′ poly(A) RNA and ssDNA exonuclease that is the catalytic component of the cytoplasmic deadenylase
Several lines of evidence indicate that the CCR4 protein possesses 3′–5′ poly(A) exoribonuclease and exodeoxyribonuclease activities. (i) Database searches revealed that the C-terminal region of CCR4 is homologous to ExoIII and HAP1 (APE1) of the AP endonuclease family (Figure 1A; Dlakic, 2000; Whisstock et al., 2000). The CCR4 homology extends to all five highly conserved catalytic and metal-binding motifs in this family of proteins. (ii) Our genetic analyses showed that mutation of four of these conserved, key catalytic amino acid residues abrogates CCR4 functions in vivo. (iii) Most importantly, full-length CCR4-FLAG fusion protein displayed poly(A) RNA and ssDNA exonuclease activities in vitro. (iv) The relative loss of enzymatic activity displayed by CCR4 mutant proteins corresponded to their relative functional inactivity in vivo. ccr4-1, -2 and -4 proteins were all completely inactive in our assays and non-functional in vivo. ccr4-3, in contrast, retained some RNA exonuclease activity and, when overexpressed in yeast, could partially complement a ccr4 deletion. Importantly, each of these ccr4 alleles reduced the rate of degradation of several mRNAs in vivo. (v) CCR4 has been shown to be a component of the cytoplasmic deadenylase, and CCR4 is required for the activity of this complex (Tucker et al., 2001). (vi) The CCR4 protein was able to display exonuclease activity as a monomer independently of the CAF1 protein. In total, our results establish that CCR4 is the catalytic component for the cytoplasmic mRNA deadenylase activity. Similar results establishing CCR4 as the catalytic subunit of the deadenylase have also been reported by Tucker et al. (2002).
The four ccr4 mutated proteins that we analyzed were non-functional in our enzymatic assays. Based on the current mechanism of action of APE1 (Mol et al., 2000), this result would imply that the catalytic mechanism of nucleotide cleavage would be conserved between CCR4 and the APE1 nuclease. The function of D780A supports this hypothesis. The D780A alteration in CCR4 resulted in a protein with some residual activity. This aspartate residue would be expected to be less important for catalysis due to its role in stabilizing the H818 residue as compared with the other residues we analyzed, which would be presumed to be involved directly in the catalytic mechanism. It also appears from the enzyme assays that CCR4 is not a processive enzyme in its degradation of poly(A) substrates. Two observations support this conclusion: a relatively homogeneous population of partially deadenylated substrates is visible and the unreacted RNA substrate is not found at the same time as fully deadenylated products (Martinez et al., 2000).
Our identification of CCR4 as an RNase defines a novel exoribonuclease superfamily in addition to the previ ously proposed six exoribonuclease families (Zuo and Deutscher, 2001). This CCR4 family of proteins most probably contains the NGL proteins in yeast and similarly related proteins from other eukaryotes, such as nocturnin and angel. The CCR4 family will probably display both RNA and ssDNA substrate preferences. This broad specificity of functions implicates this family of proteins as being potentially important to many regulatory processes, and it will require careful biochemical analysis to decipher the biological roles of each of these proteins.
Interestingly, one other member of the Mg2+-dependent ExoIII/HAP1 (APE1) family, Line L1 endonuclease, also shares a poly(A) substrate preference like that of CCR4 protein. Human L1 elements are highly abundant poly(A) (non-long terminal repeat) retroviral elements whose second open reading frame (ORF2) encodes a reverse transcriptase. The endonuclease (EN) domain at the N-terminus of ORF2 shows homology to AP endonuclease (Feng et al., 1996). However, L1 EN protein was shown to display nicking endonuclease activities on DNA that was not specific for AP-DNA substrates. The cleavage hot spots on the DNA substrate were one or more purines just 3′ to the site of cleavage, and these often involved short runs of As (Feng et al., 1996; Cost et al., 2001). Both L1 EN and CCR4 lack the AP-recognizing sequences that are found in the ExoIII and APE1 proteins. The shared poly(A) substrate preference of L1 EN and CCR4 suggests shared aspects in its recognition of the substrate, which, unfortunately, is not obvious from direct comparisons of the primary sequences of these proteins.
Roles of CAF1 and NOT proteins in mRNA degradation
In contrast to CCR4, the biochemical role of CAF1 remains less clear. It has been reported that an E.coli preparation of CAF1 contains deadenylase activity (Daugeron et al., 2001), although purified yeast FLAG-CAF1 that lacked CCR4 was inactive (Tucker et al., 2001). While the higher eukaryotic CAF1 proteins share homology to the 3′ exonuclease RNase D family (Moser et al., 1997), yeast CAF1 is missing at least three key amino acid residues considered important for this catalytic function. Mutation of two of the other putative key, conserved catalytic residues of yeast CAF1 had no effect on the ability of the protein to function in yeast (J.Chen and C.L.Denis, unpublished observations), suggesting that yeast CAF1 is not a functional member of the RNase D family. However, CAF1 may have RNA- and/or DNA-binding characteristics and thereby may help to stabilize the RNA–DNA–protein intermediates required in CCR4 enzymatic function or possibly in targeting CCR4 to its proper polynucleotide substrate. The large overlap in phenotypes between caf1 and ccr4 deletions (Draper et al., 1995; Liu et al., 1998; Denis et al., 2001) and similar effects on mRNA degradation (Tucker et al., 2001) implicate CAF1 as being integral to CCR4 function in vivo, even though it is not required for its enzymatic activity in vitro. The previous observation that high copy expression of CCR4 can complement a caf1 defect, but high copy expression of CAF1 cannot complement ccr4 (Hata et al., 1998), further suggests that CCR4 is the catalytic component of the cytoplasmic deadenylase whereas CAF1 is a regulatory component.
In addition to possible direct roles in polynucleotide recognition, CAF1 enjoys the role of linking CCR4 to the remainder of the CCR4–NOT complex. The recent evidence that several not deletions do affect mRNA degradation and the deadenylation process, albeit to a lesser extent than ccr4 and caf1 deletions (Tucker et al., 2002), suggests that the CCR4–NOT complex as a whole functions in the mRNA degradation process. CAF1 may function, therefore, to link the catalytically active end (CCR4) of the complex to other components potentially involved in RNA recognition or other aspects of the process. It may be significant in this regard that NOT4 contains a putative RNA-binding motif (Burd et al., 1994; Albert et al., 2000; our unpublished observations), deletion of which inactivates NOT4 function in vivo (J.Chen and C.L.Denis, unpublished observation). Further investigations are required to determine the role of each of the other components of the CCR4–NOT complex in the mRNA degradation process, specifically with regard to RNA recognition and regulatory aspects.
Yet, defects in the not genes do elicit phenotypes not shared by ccr4 or caf1. Most notable of these is the increased expression from a non-canonical TATAA at the HIS3 locus (Collart and Struhl, 1994; Liu et al., 1998). This effect appears to result from NOT2/NOT5 interaction with TFIID (Badarinarayana et al., 2000; Lemaire and Collart et al., 2000) and would therefore not require CCR4 nuclease function. It can be imagined that due to the distinct location of NOT2/NOT5 separate from CCR4 within the CCR4–NOT complex (Bai et al., 1999), the NOT protein end of the CCR4–NOT complex could be involved in effects on initiation, whereas the CCR4 part of the complex could be involved in other aspects of mRNA formation and degradation. This hypothesis would suggest that the CCR4–NOT complex could integrate initiation with post-initiation events, as has been suggested for the CTD of RNA polymerase II (pol II) (Hirose and Manley, 2000; Barilla et al., 2001). The observation that CCR4 is present in an RNA pol II complex (Chang et al., 1999) and that the CCR4–NOT complex components can be immunoprecipitated with factors associated with RNA pol II (Y.-C.Chiang and C.L.Denis, unpublished observation) further suggests a potentially important role for the CCR4–NOT complex in various aspects of mRNA formation.
Materials and methods
Yeast strains, growth conditions
Yeast strains used in this research are listed in Table III. The different growth conditions for genetic analysis are described in the text. Generally, yeast strains were grown at 30°C on YEP medium (2% yeast extract, 1% Bacto Peptone) or selective medium (Draper et al., 1994) supplemented with either 4% glucose or 2% galactose/2% raffinose unless otherwise indicated. YD plates consisted of YEP medium supplemented with 2% glucose and 2% agar, caffeine plates contained YD plates with 8 mM caffeine, and glycerol plates consisted of YEP medium supplemented with 3% glycerol and 2% agar.
Table III. Yeast strains.
| Strain | Genotype |
|---|---|
| MD9-7c | MATα adh1-11 his3 trp1 ura3 ccr4-10 |
| MD9-7c-120 | isogenic to MD9-7c except trp1::CCR4-TRP1 |
| MD9-7c-120-1 | isogenic to MD9-7c except trp1::ccr4-1-TRP1 |
| MD9-7c-120-2 | isogenic to MD9-7c except trp1::ccr4-2-TRP1 |
| MD9-7c-120-3 | isogenic to MD9-7c except trp1::ccr4-3-TRP1 |
| MD9-7c-120-4 | isogenic to MD9-7c except trp1::ccr4-4-TRP1 |
| KY803 | MATα leu2-PET56 trp1-1 ura3-52 gal2 gcn4-1 |
| KY803-1a-1 | isogenic to KY803 except ccr4::ura3 |
| EGY188 | MATα ura3 his3 trp1 LexA-LEU2 |
| EGY188-1a-1 | isogenic to EGY188 except ccr4::ura3 |
| EGY188-1a-1-c1 | isogenic to EGY188 except ccr4::ura3 caf1::LEU2 |
DNA plasmid constructions
Site-directed mutagenesis of the CCR4 proteins (ML400-1: E556A, ML400-2: D713A, ML400-3: D780A, ML400-4: H818A) was performed by sequential PCR amplification methods using ML400 as the template. ML400 is derived from TM10 (Malvar et al., 1992). CCR4-FLAG constructs that contain full-length CCR4 and its mutants, and which have their own endogenous promoter sequences (ML400, etc.), were first constructed by placing CCR4 sequences in pRS426. The CCR4-FLAG fusion constructs were then generated from their pRS426 derivatives by replacing their BamHI–SalI pieces at the C-terminal end with PCR-amplified BamHI-FLAG–SalI fragments (Dunckley et al., 1999). LexA–CCR4 fusions were variants of MD9 (Draper et al., 1994). Human CCR4 was constructed from human cDNA KIAA1194, a kind gift from Dr Nagase (Kazusa DNA Research Institute). The human CCR4-FLAG was constructed by linking the human CCR4 cDNA sequence directly downstream of the yeast CCR4 promoter and placing a FLAG sequence at its C-terminus. The whole human CCR4-FLAG cassette was cloned into the pRS426 vector at KpnI and SpeI sites. All the sequences of human CCR4 and yeast CCR4, as well as its derivative mutant plasmids, were verified by sequencing of both strands.
DNA and RNA substrate
The commercially synthesized RNA and ssDNA oligonucleotides used were: 3′-5As RNA, UAUGUGAAUUCUAUGCCACCCCAAAAA; 3′-20As RNA, UCUAAAUAAAAAAAAAAAAAAAAAAAAAAA; 3′-10 Cs RNA, UCUAAAUAAAAAAAAAAAAACCCCCCCCCC; 3′-5As ssDNA, TATGTGAATTCTATGCCACCCCAAAAA; 3′-CGssDNA, TATAGCATGCCTATCACATATAAATAGAGTGCCAGTAGCG; ADH2 ssDNA, GGCATACTTGATAATGAAAACTATAAATCGTAAAGACATAAG; HSP12 ssDNA, CCCTTGTTGTCTTCTGGTTGAACCTTACCAGCGACCTTGTCGG; GAL1 ssDNA, GCCCAATGCTGGTTTAGAGACGATGATAGCATTTTCTAGCTCAGCATCAGTGGATCTTAGGG; ACT1-1 ssDNA, GCGTCGTCACCGGCAAAACCGGCTTTACACATACCAGAACCG; and ACT1-2 ssDNA, CCACTTTCGTCGTATTCTTGTTTTGAGATCCACATTTGTTGGAAGGTAGTCAAAGAAGCGTCAT.
The RNA and ssDNA substrates (10 pmol) were labeled with T4 polynucleotide kinase and [γ-32P]ATP. All the labeled substrates were subjected to gel filtration on Sephadex G-25 spin columns to remove unlabeled nucleotides. The radioactive substrates were then diluted and normalized to 1 pmol/µl = 10 000 c.p.m./µl.
Protein extraction
CCR4-FLAG fusion protein purification was conducted as described previously with a few modifications (Dunckley et al., 1999). The yeast culture containing the CCR4-FLAG fusion was grown to late log phase (OD600 = 1.5) in selective medium with 4% glucose. The cells were washed and lysed in HEPES buffer (50 mM HEPES–NaOH pH 7.6, 150 mM NaCl, 2 mM MgCl2, 20% glycerol, 0.1% NP-40) plus a protease inhibitor cocktail. After clarification of the crude cell lysate by centrifugation at 14 000 g for 10 min and then ultracentrifugation at 100 000 g for 45 min at 4°C, the supernatants were incubated with 500 µl of anti-FLAG M2 affinity agarose (Sigma) at 4°C overnight. After extensive washes, the bound FLAG fusion proteins were eluted twice with HEPES buffer containing 200 µg/ml FLAG peptide (Sigma). The eluates were dialyzed against 4 l of HEPES buffer for 24 h with two changes of buffer, and concentrated using a Centricon 10 (Millipore). The concentrations of the extracts were quantified both by Bradford assay for total proteins and by comparison with a gradient of standard bovine serum albumin (BSA) concentrations for determining the CCR4-FLAG concentration. For additional Superdex 200 gel filtration chromatography purification, 0.1 mg dialyzed eluates were loaded on the Superdex 200 HR 10/30 column (Pharmacia) with HEPES running buffer without NP-40. The calibration of the Superdex 200 HR 10/30 column was: IgG (160 kDa) at fraction 24; BSA (67 kDa) at fraction 30; and β-lactoglobulin (35 kDa) at fraction 33. The CCR4-FLAG fusion protein extracts were run subsequently on an 8% SDS–polyacrylamide gel. The protein bands on the gels were analyzed either by silver staining or by standard western analysis.
In vitro RNase or DNase assay
The standard enzymatic assays (10 µl) contained HEPES binding buffer [50 mM HEPES–NaOH pH 7.6, 150 mM NaCl, 2 mM MgCl2, 10% glycerol, 1 mM dithiothreitol plus RNase inhibitors (Ambion)], 1 µl of radioactive substrate (1 pmol) and 1× protein (32 ng unless indicated otherwise in the figure legends). After incubation at room temperature, the reaction was stopped by addition of an equal volume of formamide/EDTA gel loading buffer. For the time course assays, after incubation at room temperature for the given amount of time as indicated in the figures, 10 µl of reaction mixtures were withdrawn, and the reaction was stopped by addition of an equal volume of the formamide/EDTA gel loading buffer. After boiling for 5 min, the reaction mixtures were loaded onto a 7 M urea–14% sequencing polyacrylamide denaturing gel. The products were analyzed and quantified by a phosphoimager and its attached software (Bio-Rad).
Quantitative S1 nuclease protection assay
Total RNA was obtained following standard protocols. Quantitative S1 nuclease protection assays were conducted as described (Collart and Struhl, 1993), with a few modifications. About 60 µg of total RNA were hybridized with 40 fmol of radioactively labeled oligonucleotides overnight at 55°C in hybridization buffer (40 mM HEPES pH 7.0, 500 mM NaCl, 1 mM EDTA, 1% Triton X-100). The S1 nuclease digestion reactions were carried out at 37°C for 1 h. The reaction mixtures were stopped by 5 mM EDTA and precipitated with 2 vols of ethanol. The final products were separated on a 7 M urea–10% polyacrylamide denaturing gel, and quantified by a phosphoimager and its attached software (Bio-Rad). The half-life of mRNA degradation was calculated following measurement of the ratio of the abundance of the mRNA versus the abundance of the ACT1 mRNA, as determined by densitometric analysis at each designated time point. CCR4 mutations do not affect steady-state levels of ACT1 mRNA.
Acknowledgments
Acknowledgements
We would like to thank especially R.Parker and M.Tucker for communication of results prior to publication. We also appreciate Dr Takahiro Nagase (Kazusa DNA Research Institute) for sending us the human KIAA1944 cDNA construct. The technical assistance of G.Quigley was invaluable. The research was supported by NIH grant GM41215 and HATCH project 291. This is New Hampshire Agriculture Experiment Station publication number 2111.
References
- Albert T.K., Lemaire,M., van Berkum,N.L., Gentz,R., Collart,M.A. and Timmers,H.T. (2000) Isolation and characterization of human orthologs of yeast CCR4–NOT complex subunits. Nucleic Acids Res., 28, 809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badarinarayana V., Chiang,Y.C. and Denis,C.L. (2000) Functional interaction of CCR4–NOT proteins with TATAA-binding protein (TBP) and its associated factors in yeast. Genetics, 155, 1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Salvadore,C., Chiang,Y.-C., Collart,M.A., Liu,H.-Y. and Denis,C.L. (1999) The CCR4 and CAF1 proteins of the CCR4–NOT complex are physically and functionally separate from NOT2, NOT4 and NOT5. Mol. Cell. Biol., 19, 6642–6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barilla D., Lee,B.A. and Proudfoot,N.J. (2001) Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 98, 445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzilay G., Mol,C.D., Robson,C.N., Walker,L.J., Cunningham,R.P., Tainer,J.A. and Hickson,I.D. (1995) Identification of critical active-site residues in the multifunctional human DNA repair enzyme HAP1. Nature Struct. Biol., 2, 561–568. [DOI] [PubMed] [Google Scholar]
- Burd C.G. and Dreyfuss,G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science, 265, 615–621. [DOI] [PubMed] [Google Scholar]
- Chang M., French-Cornay,D., Fan,H.-Y., Klein,H., Denis,C.L. and Jaehning,J.A. (1999) A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p and Ccr4p plays a role in protein kinase C signaling. Mol. Cell. Biol., 19, 1056–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Rappsilber,J., Chiang,Y.-C., Russell,P., Mann,M. and Denis, C.L. (2001) Purification and characterization of the 1.0 MDa CCR4–NOT complex identifies two novel components of the complex. J. Mol. Biol., 314, 683–694. [DOI] [PubMed] [Google Scholar]
- Collart M.A. and Struhl,K. (1993) CDC39, an essential nuclear protein that negatively regulates transcription and differentially affects the constitutive and inducible HIS3 promoters [published erratum appears in EMBO J. (1993), 12, 2990]. EMBO J., 12, 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart M.A. and Struhl,K. (1994) NOT1(CDC39), NOT2(CDC36), NOT3 and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev., 8, 525–537. [DOI] [PubMed] [Google Scholar]
- Cost G.J., Golding,A., Schlissel,M.S. and Boeke,J.D. (2001) Target DNA chromatinization modulates nicking by L1 endonuclease. Nucleic Acids Res., 29, 573–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugeron M.-C., Mauxion,F. and Seraphin,B. (2001) The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res., 29, 2448–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis C.L. (1984) Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics, 108, 833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis C.L. and Malvar,T. (1990) The CCR4 gene from Saccharomyces cerevisiae is required for both nonfermentative and spt-mediated gene expression. Genetics, 124, 283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis C.L., Chiang,Y.-C., Cui,Y. and Chen,J. (2001) Genetic evidence supports a role for the yeast CCR4–NOT complex in transcriptional elongation. Genetics, 158, 627–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlakic M. (2000) Functionally unrelated signaling proteins contain a fold similar to Mg2+-dependent endonucleases. Trends Biochem. Sci., 25, 272–273. [DOI] [PubMed] [Google Scholar]
- Draper M.P., Liu,H.-Y., Nelsbach,A.H., Mosley,S.P. and Denis,C.L. (1994) CCR4 is a glucose-regulated transcription factor whose leucine-rich repeat binds several proteins important for placing CCR4 in its proper promoter context. Mol. Cell. Biol., 14, 4522–4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper M.P., Salvadore,C. and Denis,C.L. (1995) Identification of a mouse protein whose homolog in Saccharomyces cerevisiae is a component of the CCR4 transcriptional regulatory complex. Mol. Cell. Biol., 15, 3487–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley T. and Parker,R. (1999) The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J., 18, 5411–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A., Barbot,W., Loireau,M.P. and Heidmann,T. (1999) Characterization of a mammalian gene related to the yeast CCR4 general transcription factor and revealed by transposon insertion. J. Biol. Chem., 274, 31068–31075 [DOI] [PubMed] [Google Scholar]
- Feng Q., Moran,J.V., Kazazian,H.H.,Jr and Boeke,J.D. (1996) Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell, 87, 905–916. [DOI] [PubMed] [Google Scholar]
- Gorman M.A., Morera,S., Rothwell,D.G., de La Fortelle,E., Mol,C.D. Tainer,J.A., Hickson,I.D. and Freemont,P.S. (1997) The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J., 16, 6548–6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C.B. and Besharse,J. (1996) Identification of a novel vertebrate circadian clock regulated gene encoding the protein nocturnin. Proc. Natl Acad. Sci. USA, 93, 14884–14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata H., Mitsui,H., Liu,H.-Y., Bai,Y., Denis,C.L., Shimizu,Y. and Sakai,A. (1998) Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics, 148, 571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y.J. and Manley,L. (2000) RNA polymerase II and the integration of nuclear events. Genes Dev., 14, 1415–1529. [PubMed] [Google Scholar]
- Kurzik-Dumke U. and Zengerle,A. (1996) Identification of a novel Drosophila melanogaster gene, angel, a member of a nested gene cluster at locus 59F4,5. Biochim. Biophys. Acta, 1308, 177–181. [DOI] [PubMed] [Google Scholar]
- Lemaire M. and Collart,M.A. (2000) The TATA-binding protein-associated factor yTafII19p functionally interacts with components of the global transcriptional regulator Ccr4–Not complex and physically interacts with the Not5 subunit. J. Biol. Chem., 275, 26925–2634. [DOI] [PubMed] [Google Scholar]
- Liu H.-Y., Toyn,J.H., Chiang,Y.-C., Draper,M.P., Johnston,L.H. and Denis,C.L. (1997) DBF2, a cell cycle-regulated protein kinase, is physically and functionally associated with the CCR4 transcriptional regulatory complex. EMBO J., 16, 5289–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.-Y., Badarinarayana,V., Audino,D.C., Rappsilber,J., Mann,M. and Denis,C.L. (1998) The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J., 17, 1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.-Y. et al. (2001) Characterization of CAF4 and CAF16 reveals a functional connection between the CCR4–NOT complex and a subset of SRB proteins of the RNA polymerase II holoenzyme. J. Biol. Chem., 276, 7541–7548. [DOI] [PubMed] [Google Scholar]
- Lucas J.A., Masuda,Y., Bennett,R.A., Strauss,N.S. and Strauss,P.R. (1999) Single-turnover analysis of mutant human apurinic/apyrimidinic endonuclease. Biochemistry, 38, 4958–4964. [DOI] [PubMed] [Google Scholar]
- Malvar T., Biron,R., Kaback,W. and Denis,C.L. (1992) The CCR4 protein from Saccharomyces cerevisiae contains a leucine-rich repeat region which is required for its control of ADH2 gene expression. Genetics, 132, 951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J., Ren,Y.-G., Thuresson,A.-C., Hellman,U., Astrom,J. and Virtanen,A. (2000) A 54 kDa fragment of the poly(A)-specific ribonuclease is an oligomeric, processive and cap-interacting poly(A)-specific 3′-exonuclease. J. Biol. Chem., 275, 24222–24230. [DOI] [PubMed] [Google Scholar]
- Mol C.D., Kuo,C.F., Thayer,M.M., Cunningham,R.P. and Tainer,J.A. (1995) Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature, 374, 381–386. [DOI] [PubMed] [Google Scholar]
- Mol C.D., Izumi,T., Mitra,S. and Trainer,J.A. (2000) DNA-bound structures and mutants reveal abasic DNA binding by APE1 DNA repair and coordination. Nature, 403, 451–456. [DOI] [PubMed] [Google Scholar]
- Moser M.J., Holley,W.R., Chatterjee,A. and Mian,I.S. (1997) The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res., 25, 5110–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.H., Barsky,D., Erzberger,J.P. and Wilson,D.M.,III (2000) Mapping the protein–DNA interface and the metal-binding site of the major human apurinic/apyrimidinic endonuclease. J. Mol. Biol., 298, 447–459. [DOI] [PubMed] [Google Scholar]
- Sakai A., Chibazakura,T., Shimizu,Y. and Hishinuma,F. (1992) Molecular analysis of POP2 gene, a gene required for glucose-derepression of gene expression in Saccharomyces cerevisiae. Nucleic Acids Res., 20, 6227–6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M., Valencia-Sanchez,M.A., Staples,R.R., Chen,J., Denis,C.L. and Parker,R. (2001) The transcription factor associated proteins, Ccr4p and Caf1p, are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell, 104, 377–386. [DOI] [PubMed] [Google Scholar]
- Tucker M., Staples,R.R., Valencia-Sanchez,M.A., Muhlrad,D. and Parker,R. (2002) Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J., 21, 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisstock J.C., Romero,S., Gurung,R., Nandurkar,H., Ooms,L.M., Bottomley,S.P. and Mitchell,C.A. (2000) The inositol polyphosphate 5-phosphatases and the apurinic/apyrimidinic base excision repair endonucleases share a common mechanism for catalysis. J. Biol. Chem., 275, 37055–37061. [DOI] [PubMed] [Google Scholar]
- Wilson D.M. III, Takeshita,M. and Demple,B. (1997) Abasic site binding by the human apurinic endonuclease, Ape and determination of the DNA contact sites. Nucleic Acids Res., 25, 933–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y. and Deutscher,M. (2001) Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res., 29, 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]