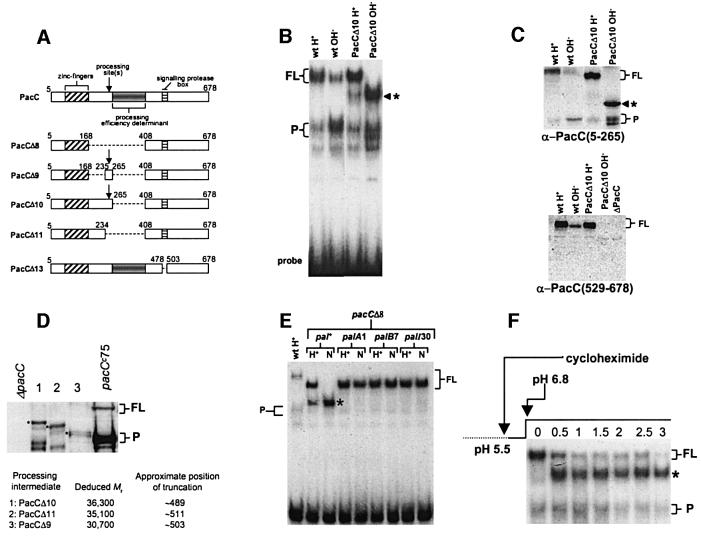
Fig. 1. (A) Schematic representation of PacC deletion mutations used in this work. To avoid confusion with earlier publications, numbering indicates amino acid positions on the basis of 678 residues, although translation starts at codon 5 (Mingot et al., 1999). (B) A processing intermediate is detected with PacCΔ10. EMSA shows PacC–DNA complexes corresponding to the different PacC forms. Strains and growth conditions are indicated. Here, and in all other panels, an asterisk indicates the processing intermediate; FL and P indicate full-length and processed form complexes, respectively. (C) Western blot analysis of protein extracts used in (B). Proteins were detected with the indicated antisera. The anti-PacC(529–678) antiserum was raised against the C-terminal PacC domain. (D) Deducing the approximate C-terminal residue of the processing intermediate. Protein extracts corresponding to the indicated mutant proteins were analysed by western blot with α-PacC(5–265) antiserum. The deduced molecular weight of the intermediates was used to estimate the approximate position of their C-terminal truncations. The pacCc75 translation product (Mr 45 478) was used as internal standard. (E) pH and pal dependence of the formation of the processing intermediate in pacCΔ8 strains. Protein extracts from acidic (H+) and neutral (N) growth conditions were analysed by EMSA. Neutral (pH 6.8) conditions leading to pH signalling were used as strains carrying palA1 or palB7 loss-of-function mutations do not grow at alkaline pH, and palI30 mutants grow poorly. (F) The translation product of PacCΔ10 and the processing intermediate show a precursor–product relationship. The relevant portion of an EMSA gel showing conversion of the PacCΔ10 translation product to the processing intermediate. Cells were grown under acidic conditions, incubated in the presence of cycloheximide for 30 min and shifted to neutral conditions also in the presence of cycloheximide. Numbers indicate hours after the pH shift.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.