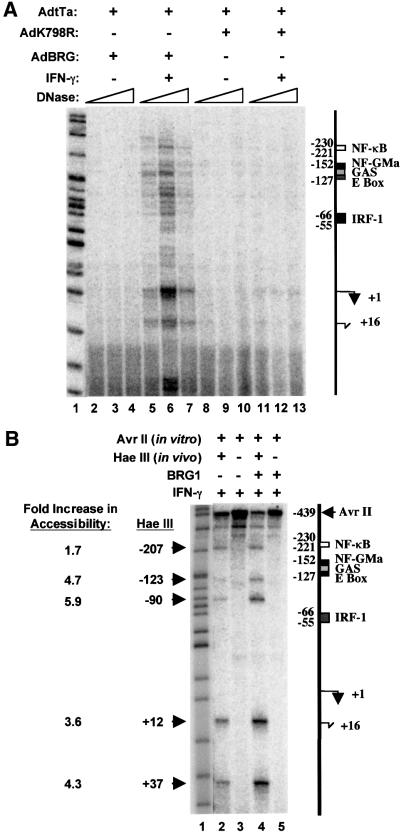
Fig. 5. BRG1-dependent chromatin remodeling at the CIITA promoter. (A) DNase I accessibility requires BRG1. SW13 cells were infected with AdtTa plus AdBRG (lanes 2–7), or AdK798R (lanes 8–13). After 20 h, cells were left untreated (lanes 2–4 and 8–10) or exposed to IFN-γ for 24 h (lanes 5–7 and 11–13). Nuclei were harvested and treated with 12.8 (lanes 2, 5, 8 and 11), 25.6 (lanes 3, 6, 9 and 12) or 51.2 (lanes 4, 7, 10 and 13) ng/µl DNase I for 3 min at 37°C. DNA was isolated and analyzed by LM-PCR. (B) HaeIII accessibility requires BRG1. Uninfected SW13 cells (lanes 2 and 3) or cells infected with AdtTa and AdBRG (lanes 4 and 5) were treated for 24 h with IFN-γ (lanes 2–5). Nuclei were isolated and incubated in vivo with HaeIII (lanes 2 and 4) or no enzyme (lanes 3 and 5) at 37°C for 10 min (lanes 2 and 4). DNA was extracted, digested to completion in vitro with AvrII and analyzed by LM-PCR. The intensity of each HaeIII band in lanes 2 and 4 was normalized to that of the full-length primer extension product to the AvrII site. The ratio of these values gave the fold increase in HaeIII accessibility. Arrows on the left and right indicate the HaeIII and AvrII sites, respectively. The marker in both (A) and (B) (lane 1) is MspI-digested pBR322 DNA. The position of elements in the CIITA promoter is indicated to the right of each figure. Major (+1) and minor (+16) start sites of transcription are indicated.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.