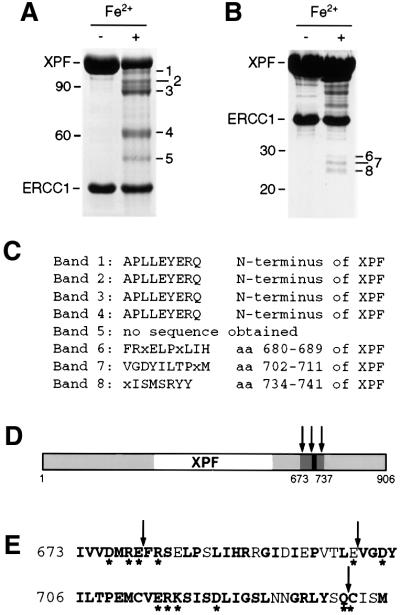
Fig. 2. Iron-induced cleavage of XPF–ERCC1. (A) SDS–PAGE gel (8%) of purified XPF–ERCC1 before and after treatment with 50 µM FeCl2 and 20 mM ascorbate for 15 h at 0°C. The five observed XPF cleavage products were designated bands 1–5, in order of decreasing size and with estimated molecular weights of 102, 95, 88, 62 and 52 kDa, respectively. A 10 kDa protein ladder (Life Technologies) was used as a marker. The positions of the 90 and 60 kDa marker bands are indicated. (B) SDS–PAGE gel (12%) of XPF–ERCC1 treated as in (A). The three bands between 25 and 30 kDa were designated bands 6–8. The positions of the 30 and 20 kDa marker bands are indicated. (C) Results from the N-terminal sequencing of the peptide fragments from bands 1–8 in (A) and (B). (D) Location of the cleavage sites in XPF. Regions of XPF that are conserved between species are shown in gray, the region encompassing the cleavage sites in dark gray and the highly conserved V/IERKX3D motif in black. The cleavage sites are indicated by arrows. (E) The amino acid sequence of region 673–737 of XPF. Conserved residues are in bold, residues selected for site- directed mutagenesis are marked with an asterisk and cleavage sites are marked with arrows.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.