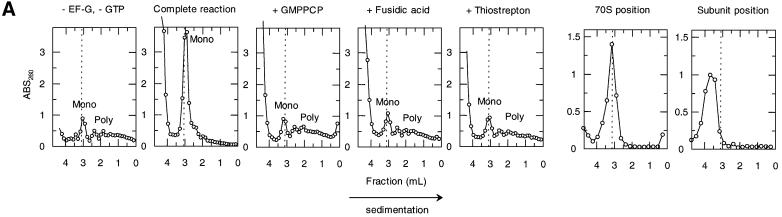
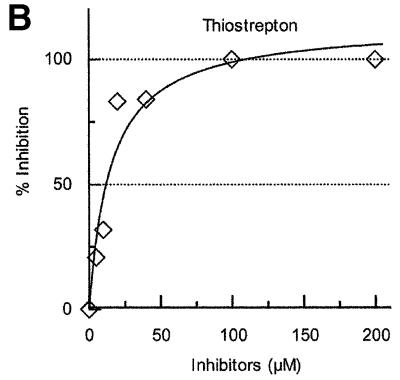
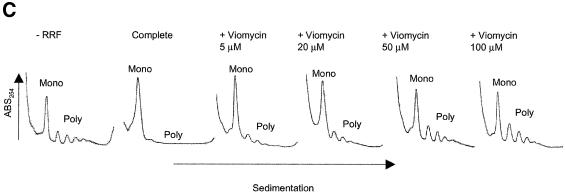
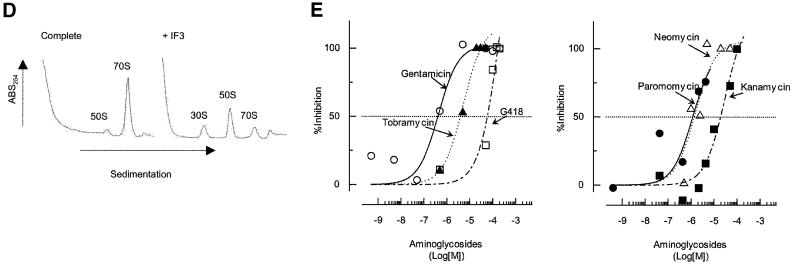
Fig. 3. Effects of antibiotics on the release of mRNA from ribosome by RRF and EF-G. (A) Effects of fusidic acid, GMPPCP (EF-G release inhibitors) and thiostrepton. The release of ribosome from the model substrate was examined by measuring the conversion of puromycin-treated polysome (marked as Poly) to monosome (marked as Mono) as described in Materials and methods except that 760 pmol of RRF and EF-G, and 3.2 A260 units of polysome were used. The ribosome sedimentation pattern was analyzed by 15–30% sucrose density gradient ultracentrifugation (Beckman SW50.1, 40 000 r.p.m., 75 min). Fractions were taken from the bottom of the tube and the A260 was measured. Sedimentation is from left to right. The first and second panel from the left show the control (minus EF-G) and complete reaction mixture, respectively. Note that in the control, the major portion of the ribosomes were polysomes, while the complete mixture showed that the polysomes were converted to the monosome. The effect of 370 µM GMPPCP (third panel from the left), 200 µM fusidic acid (fourth panel) and 20 µM thiostrepton (fifth panel) are shown. In the experiment where the effect of GMPPCP was tested, GTP was omitted. The positions of 70S ribosomes (6th panel) and subunits (7th panel) were determined by sedimentation of washed ribosome (2.0 A260 units) either in buffer R or in buffer R containing 8 mM EDTA, respectively. Note that the background level of these panels was completely flat, indicating that the polysomes observed in the first, third, fourth and fifth panels were significantly above the background. (B) Effect of thiostrepton. Effect of various amounts of thiostrepton on the release of mRNA from the model post-termination complex was examined as in (A). (C) Effect of various concentration of viomycin. Experimental conditions were the same as (A) except that A254 was measured continuously by an ISCO UA-6 spectrophotometer (ISCO). Sedimentation was from left to right. (D) Absence of ribosome subunit formation during the RRF reaction—effect of IF3. Left trace: the experimental conditions were identical to those of (C) except that the ultracentrifugation time was increased to 240 min. Right trace: the same as the left panel except that 100 µg of IF3 was added. (E) Effect of aminoglycosides. The conditions were the same as (B) except that an ISCO UA-6 spectrophotometer was used. Gentamicin (open circles), tobramycin (closed triangles), G418 (open squares), paromomycin (closed circles), neomycin (open triangles) and kanamycin (closed squares) were tested. In (B) and (E), 88% of polysomes were converted to monosome by RRF and EF-G without inhibitor, and this value was set as 100% conversion.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.