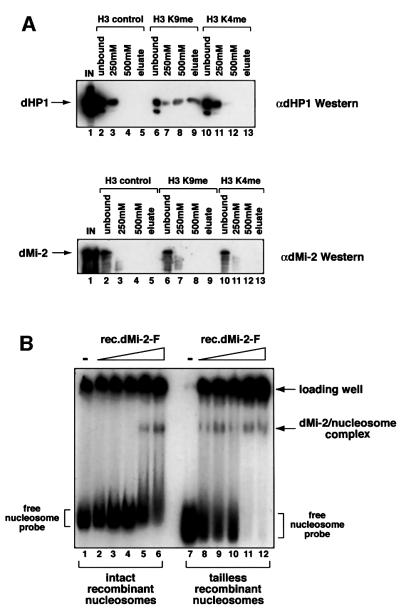
Fig. 4. dMi-2 does not require histone tails for nucleosome binding. (A) Drosophila embryo nuclear extract was incubated with immobilized methylated and unmethylated histone H3 peptides as indicated. Peptide beads were successively washed with buffer containing increasing concentration of NaCl and eluted with glycine buffer pH 2.5, as indicated on top of the panels. Proteins in unbound, wash and eluate fractions were detected by western analysis using antisera directed against dHP1 (upper panel) and dMi-2 (lower panel). IN, input; control, unmodified peptide; K9me, K9-methylated peptide; K4me, K4-methylated peptide. (B) dMi-2 nucleosome bandshift assay using a 146 bp nucleosome probe reconstituted from intact recombinant histones (lanes 1–6) or reconstituted from tailless recombinant histones (lanes 7–12). Increasing amounts of recombinant dMi-2 WT (lanes 2 and 8, 50 fmol; lanes 3 and 9, 100 fmol; lanes 4 and 10, 200 fmol; lanes 5 and 11, 300 fmol; lanes 6 and 12, 400 fmol) were incubated with radioactively labelled nucleosome probe. Complexes were resolved by native PAGE. The positions of free nucleosome probe, dMi-2–nucleosome complexes and nucleosome probe precipitated in the loading well are indicated.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.