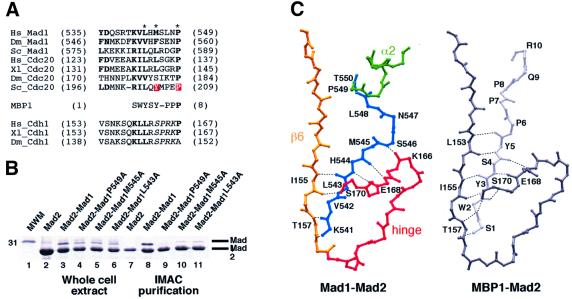
Fig. 3. Interaction of Mad2 with Mad2 binding motifs. (A) Mad2 binding motifs in Mad1, Cdc20 and MBP1. Dm: Drosophila melanogaster, other abbreviations as in Figure 2. The motif is also present in CDH1, and contains a potential CDK or MAPK target site (italics). Red boxes indicate mutation sites affecting Cdc20–Mad2 and the asterisks Mad1 mutations analysed in our study. (B) His-Mad2–Mad1485–718 and indicated mutants were co-expressed in bacteria, IMAC-purified, and analysed by SDS–PAGE. Lane 1, molecular weight marker; lanes 2–6, total bacterial lysates of indicated expression tests, showing overall protein production; lanes 7–11, IMAC-purified samples. (C) Comparison of Mad1 and MBP1 binding to Mad2. Mad1–Mad2 is shown with the colour scheme of Figure 2B. Most structural elements were removed to improve readability, and only main chain atoms are shown. Equivalent region of MBP1 (light grey) and Mad2 (dark grey) are also shown. Hydrogen bonds are shown as dashed lines. Ligand and Mad2 residues involved in main chain hydrogen bonding are labelled.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.