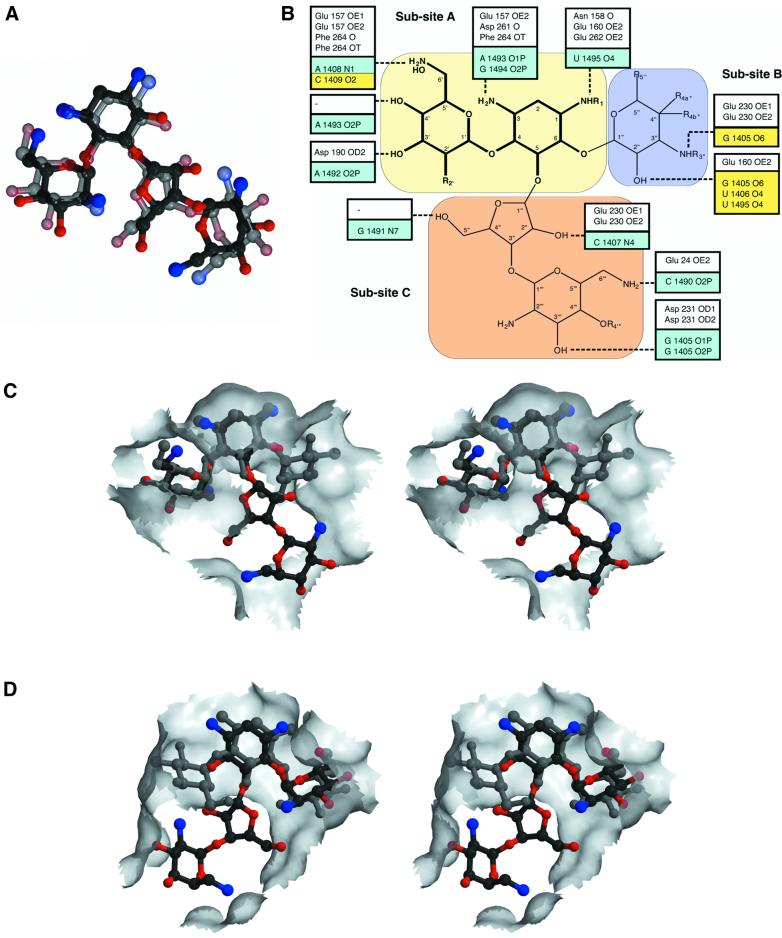
Fig. 4. Comparison of aminoglycoside binding to APH(3′)-IIIa versus the bacterial ribosome. (A) Superposition of neomycin B, in the conformation observed in the APH(3′)-IIIa ternary complex, and paromomycin I in the conformation observed in the crystal structure of the 30S ribosomal subunit (Carter et al., 2000). Neomycin B is shown in solid colors and paromomycin I is semi-transparent. (B) Schematic overview of hydrogen bond interactions made by aminoglycosides with APH(3′)-IIIa and the bacterial ribosome (Fourmy et al., 1998; Yoshizawa et al., 1998; Carter et al., 2000). A combined generic chemical structure for 4,6- and 4,5-disubstituted aminoglycosides is shown, highlighting common functional groups (see also Table I). Hydrogen bond interactions made by APH(3′)-IIIa with aminoglycosides are shown in white boxes, while those made by the ribosome, as observed in the ribosome–paromomycin I crystal structure (Carter et al., 2000), are shown in aqua. Additional interactions made by the ribosome, as observed in NMR studies of neomycin B and gentamycin C1a (Fourmy et al., 1998; Yoshizawa et al., 1998), are displayed in yellow boxes. (C) Stereo view of the van der Waals surface of the APH(3′)-IIIa aminoglycoside-binding pocket. Also shown are kanamycin A and neomycin B. (D) Stereo view of the van der Waals surface of the bacterial ribosomal aminoglycoside-binding pocket. Also shown are paromomycin I and a modeled kanamycin A. This stereo view has been subjected to a 180° rotation around the vertical axis with respect to (C), to show that opposite faces of the aminoglycosides form predominant van der Waals interactions with either the ribosome or APH(3′)-IIIa.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.