Abstract
PA28 is a γ-interferon-induced complex that associates with the 20S proteasome and stimulates breakdown of small peptides. Recent immunoprecipitation studies indicate that, in vivo, PA28 also exists in larger complexes that also contain the 19S particle, which is required for ATP-ubiquitin-dependent degradation of proteins. However, because of its lability, the structure and properties of this larger complex remain unclear. Here, we demonstrate that, in vitro, PA28 can associate with ‘singly capped’ 26S (i.e. 19S–20S) proteasomes. Electron microscopy of the resulting structures revealed one PA28 ring at one end of the 20S particle and a 19S complex at the other. These hybrid complexes show enhanced hydrolysis of small peptides, but no significant increase in rates of protein breakdown. Nevertheless, during breakdown of proteins, the complexes containing PA28αβ or PA28α generated a pattern of peptides different from those generated by 26S proteasomes, without altering mean product length. Presumably, this change in peptides produced accounts for the capacity of PA28 to enhance antigen presentation.
Keywords: antigen processing/MHC class I/PA28/proteasomes/protein degradation
Introduction
PA28 (also termed REG) is a ring-shaped 11S (180 kDa) multimeric complex that can bind to the two ends of the 20S proteasome and dramatically stimulate its capacity to hydrolyze small peptides (Ma et al., 1992). In mammals, PA28 is composed of two homologous subunits, PA28α and PA28β, both of which are induced by γ-interferon (Rechsteiner et al., 2000), which is a potent stimulator of major histocompatibility complex (MHC) class I antigen presentation (Rock and Goldberg, 1999). This complex has been reported by one group (Ahn et al., 1996) to be hexameric (3α3β) and by others to be a heptamer (3α4β) (Zhang et al., 1999). By itself, PA28α in vitro forms a heptameric ring (Knowlton et al., 1997) that can stimulate peptide hydrolysis by 20S particles to the same extent as the heteromeric (3α3β) complex (Song et al., 1996).
Most peptides presented on surface MHC molecules are generated by proteasomes during the course of intracellular protein degradation (Rock et al., 1994). Amongst its multiple actions, γ-interferon induces alternative forms of the 20S proteasome (‘immunoproteasomes’) with distinct β-subunits (Rock and Goldberg, 1999), which alter its cleavage specificity so as to enhance the production of antigenic peptides (Cascio et al., 2001). PA28, therefore, is assumed to promote antigen presentation, and transfection of PA28α alone has been reported to enhance class I presentation of some (Groettrup et al., 1996), but not all, antigens (Schwarz et al., 2000). However, other investigators have failed to demonstrate such an effect (Rock and Goldberg, 1999).
The precise role of PA28αβ and its homologs, PA28γ in mammals (also called the Ki antigen) and PA26 in trypanosomes, remains unclear and controversial (Rock and Goldberg, 1999; Rechsteiner et al., 2000). Mammalian PA28 was first isolated by DeMartino’s (Ma et al., 1992) and Rechsteiner’s (Dubiel et al., 1992) laboratories as an activator of peptide hydrolysis by the 20S particle, and a variety of biochemical actions have been proposed for this complex, including allosteric activation of the proteasome’s active sites (Ma et al., 1992), stimulation of peptide entry into the 20S particle (Ma et al., 1992), stimulation of peptide exit (Whitby et al., 2000) and facilitating the binding of proteasomes to chaperones or to components of the endoplasmic reticulum (Rechsteiner et al., 2000). PA28 has also been reported (Preckel et al., 1999) to be essential for the assembly of immunoproteasomes, the alternative forms induced by γ-interferon, but subsequent studies have failed to confirm this finding (Murata et al., 2001). Deletion of PA28 genes also causes a reduced capacity to present certain antigens and not others (Rock and Goldberg, 1999). Two groups have reported that, in vitro, PA28 enhances the capacity of 20S proteasomes to make both the correct C- and N-terminal cleavages in longer oligopeptides that convert them to class I-presented epitopes (Dick et al., 1996; Shimbara et al., 1997). Consequently, it was proposed that PA28 promotes retention of peptide intermediates within the proteolytic chamber so as to favor such double cleavages (Dick et al., 1996; Shimbara et al., 1997).
Recent studies, however, indicate that proteasomes generate the C-terminal residues of antigenic peptides, while their N-termini can be produced primarily by trimming of proteasome products by intracellular aminopeptidases (Cascio et al., 2001), a process also stimulated by γ-interferon (Beninga et al., 1998). Moreover, recently, the crystal structure of PA26 (the PA28 homolog) in association with the 20S yeast particle has been solved by X-ray diffraction by Hill and co-workers (Whitby et al., 2000). In this structure, the binding of PA26 opens the normally closed central channel in the proteasome’s α-ring, through which substrates enter the degradative particle (Groll et al., 2000), and products exit (Kohler et al., 2001). On this basis, PA28 was predicted to cause the release of peptide products of greater mean length. In fact, yeast proteasomes with a mutation that prevents closing of this gate do yield peptide products of greater mean size than normal particles (Kohler et al., 2001). Most (70%) proteasomal products are shorter than eight residues in length (Kisselev et al., 1999), the minimum size needed for binding to MHC molecules. Therefore, premature release of peptides from the proteasome would be expected to enhance the fraction of products capable of serving in antigen presentation, either directly or after trimming by aminopeptidases in the cytosol (Mo et al., 2000; Stoltze et al., 2000) or endoplasmic reticulum (Snyder et al., 1994; Craiu et al., 1997).
Although, in vitro, PA28 stimulates proteasomal degradation of short peptides, it seems unlikely that PA28 functions in vivo to stimulate hydrolysis of cytosolic oligopeptides within proteasomes because such peptides are hydrolyzed very rapidly by other cytosolic peptidases (Saric et al., 2001; T.Saric and A.L.Goldberg, in preparation). Moreover, PA28 by itself cannot support the ATP-dependent hydrolysis of proteins or ubiquitin-conjugated proteins by the proteasome (Dubiel et al., 1992; Ma et al., 1992; Kuehn and Dahlmann, 1996). Protein degradation by the ubiquitin–proteasome pathway requires the 26S proteasome complex, which is composed of the 20S core particle capped at one or both of its ends by 19S regulatory particles (Voges et al., 1999). Unlike PA28, the 19S complex contains binding sites for ubiquitin chains (Thrower et al., 2000) and six ATPase subunits that catalyze protein unfolding and translocation into the 20S core particle (Braun et al., 1999; Benaroudj and Goldberg, 2000; Navon and Goldberg, 2001).
Of particular interest was the recent finding by Tanaka, Hendil and co-workers that immunoprecipitates of 19S particles from cells contain not only 20S proteasomes, but also the PA28 complex (Hendil et al., 1998). These investigators further showed that γ-interferon treatment of cells led to the appearance of these hybrid complexes (Tanahashi et al., 2000). However, these novel structures are very labile (Tanahashi et al., 2000), and attempts by multiple investigators to isolate them have been unsuccessful. Because they do not withstand standard chromatographic methods or exposure to high ionic strength buffers (Tanahashi et al., 2000), their structural organization, functional properties and in vivo significance have remained uncertain.
In the present study, we have avoided this problem of instability by reconstituting these hybrid complexes in vitro from highly purified 26S proteasomes and recombinant PA28α or native PA28 containing both the α and the β subunits. After this manuscript was submitted for publication, Kopp et al., (2001) reported similar in vitro reconstitution of these hybrid complexes using PA28αβ but did not investigate their capacity for protein degradation. In the present study, we have used the reconstituted PA28–20S–19S complexes to investigate their structure, their capacity to degrade proteins and the nature of the peptide products generated. These studies have succeeded in defining several unexpected biochemical features of these hybrid complexes that appear likely to be of importance in the stimulation of antigen presentation.
Results
In order to study the properties of complexes containing PA28 and 26S proteasomes, we initially purified recombinant mouse PA28α (Ahn et al., 1995) expressed in Escherichia coli and 20S and 26S proteasomes from rabbit spleen, as described previously (Cascio et al., 2001). Spleen was chosen initially because it contains exclusively immunoproteasomes (Van Kaer et al., 1994; Eleuteri et al., 1997; Cascio et al., 2001), the specialized form induced together with PA28 by γ-interferon (Realini et al., 1994). The 26S proteasomes were purified to apparent homogeneity, although, as found by native PAGE, these preparations consisted of approximately equal amounts of a mixture of singly capped complexes (19S–20S) and doubly capped symmetric complexes (19S–20S–19S) (Figure 1B). As expected, purified PA28α, like natural PA28αβ (see below), dramatically stimulated all three peptidase activities of the 20S particles, as measured with fluorogenic substrates specific for the chymotrypsin-like (Suc-LLVY-amc), trypsin-like (Boc-LLR-amc) and caspase-like (post-acidic) active site (Z-YVAD-amc) (not shown).
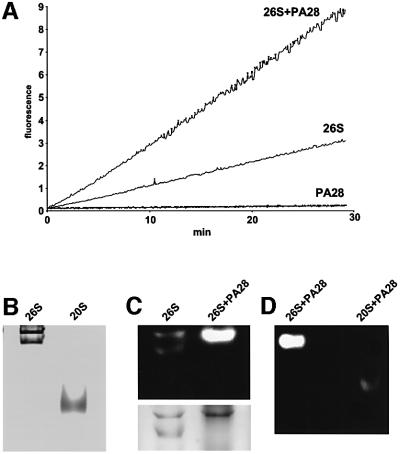
Fig. 1. PA28 interacts with 26S proteasomes and stimulates peptide hydrolysis. (A) 26S proteasomes (10 pM) were pre-incubated in 20 mM Tris–HCl pH 7.5, 2 mM ATP, 5 mM MgCl2, 1 mM EDTA at 37°C with and without PA28 (20 nM). Reactions were started by adding Suc-LLVY-amc at a final concentration of 100 µM, and the fluorescence of released amc was measured. (B) Native PAGE of 26S and 20S proteasomes. Note the two bands corresponding to singly and doubly capped 26S. (C) A 5 µg aliquot of 26S proteasomes was pre-incubated in the absence and presence of 3 µg of PA28 and separated by native PAGE. The gel was developed by fluorogenic peptide overlay, and the same gel was then stained with Coomassie Blue. (D) After association with PA28, 26S and 20S migrate in completely different positions. A 5 µg aliquot of 26S and 1 µg of 20S were pre-incubated with 3 µg of PA28 and then subjected to native PAGE.
Activation of singly capped 26S proteasomes by PA28
Surprisingly, PA28 was also found to increase 3- to 4-fold the activity of the 26S proteasomes against Suc-LLVY-amc, the substrate of the chymotrypsin-like active sites (Figure 1A). A similar 3- to 4-fold activation was also found with peptide substrates specific for the trypsin- and caspase-like sites of the proteasome (data not shown). This enhancement of the peptidase activity of the 26S proteasome was not seen in the presence of 20 mM NaCl (data not shown), which can block PA28 binding to the 20S particle (Chu-Ping et al., 1994; Kuehn and Dahlmann, 1996). In similar experiments using native PA28αβ in place of the recombinant PA28α, a very similar 3- to 4-fold activation of peptide hydrolysis by 26S proteasomes was seen (data not shown). However, this stimulation of the 26S proteasomes was obtained with significantly lower concentrations of the native molecule than of the recombinant complex, which lack the β-subunits (i.e. the Kd for this interaction was 2 nM with PA28αβ and 10 nM with PA28α).
This stimulation of peptidase activity of the 26S proteasomes was unexpected (Chu-Ping et al., 1994; Hoffman and Rechsteiner, 1994) and was not due to an activation of contaminating 20S particles, since no such particles could be detected in our 26S preparations by gel electrophoresis (Figure 1B), even after several hours incubation (Figure 3B). In addition, upon native gel electrophoresis, no activity was seen in the region of the gel that corresponds to the PA28–20S–PA28 complexes (Figure 1D) studied by previous workers (Dubiel et al., 1992; Hoffman and Rechsteiner, 1994; Realini et al., 1994). Moreover, after incubation with PA28, the migration of the 26S preparation upon native PAGE changed dramatically due to formation of a novel hybrid particle at the expense of the singly capped 19S–20S complexes. Specifically, the band identified by Coomassie Blue staining and peptidase activity as the singly capped form disappeared and, simultaneously, there was a several-fold increase in the peptidase activity and Coomassie Blue staining of a broad higher molecular weight band, which included the doubly capped form (Figure 1C). Thus, the greater peptidase activity found in Figure 1A was associated with PA28-induced modification and activation of all, or nearly all, the singly capped forms (Figure 1C).
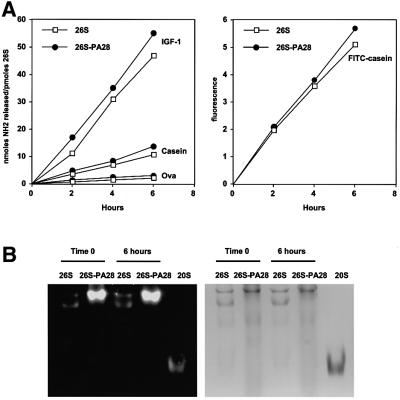
Fig. 3. (A) Association of PA28 with the 26S proteasomes does not increase the rate of protein breakdown. Degradation of reductively methylated IGF-1 (1.5 mM), casein (1 mM), ovalbumin (10 µM) and FITC–casein (10 µM) was performed at 37°C in 20 mM Bis-Tris-propane pH 7.5, 2 mM ATP, 5 mM MgCl2, 1 mM EDTA with a concentration of the enzyme variable between 10 and 30 nM depending on the substrate. At the indicated times, aliquots were removed and analyzed for the appearance of amino groups with fluorescamine (for IGF-1, casein and ovalbumin) or for the generation of soluble fluorescence after precipitation with perchloric acid (for FITC–casein). (B) Even after a 6 h incubation at 37°C, only 26S and hybrid complexes could be detected in the digestion assays. 26S and 26S–PA28 were incubated for 6 h in the presence of casein. At time 0 and after 6 h, an aliquot was removed and loaded onto a native polyacrylamide gel. The gel was overlaid with fluorogenic substrate, and then Coomassie Blue stained. Even after 6 h, 26S particles (5 µg) in the presence of PA28 were several fold more active than control 26S alone, and no 20S particles could be detected.
Although these initial experiments utilized 26S immunoproteasomes isolated from spleen, which contain only the γ-interferon-induced β-subunits (Van Kaer et al., 1994; Eleuteri et al., 1997; Cascio et al., 2001), subsequent experiments with 26S normal proteasomes isolated from rabbit muscle yielded very similar results upon addition of PA28α. In addition, a similar stimulation of 26S proteasomes was seen in analogous experiments upon addition of the heteromeric complex PA28αβ (see below).
Electron microscope analysis of hybrid particles
To investigate further the structure of this new ‘hybrid’ form of proteasomes, we analyzed our 26S samples by negative stain electron microscopy (EM), in the presence and absence of PA28. As expected, images taken from preparations of 26S proteasomes showed 20S core particles capped at one or both ends by 19S particles in equal proportions, although, after dilution and staining, ∼5% of the particles appeared to be uncapped 20S proteasomes (data not shown). These sharp images of 26S complexes are consistent with those obtained by previous investigators (Adams et al., 1997; Walz et al., 1998).
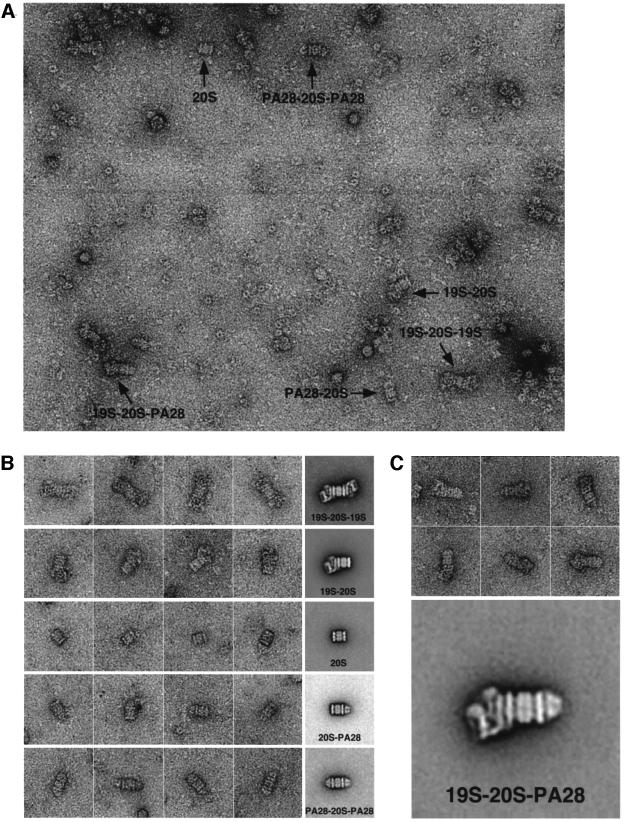
However, the negatively stained images of 26S proteasomes that were incubated with PA28 were quite different (Figure 2A). A major new species was found: a hybrid complex composed of a 20S proteasome bound at one end to a 19S particle and at the other to a PA28 ring. In addition, we also observed some proteasomes that were singly or doubly capped with PA28 rings. We recorded >130 images of such PA28-containing preparations, from which we selected ∼4600 particles. The particles on the grid were grouped manually into six classes; the particles of each class were subjected to multireference alignment, and representative class averages are shown in Figure 2B, along with a gallery of particles that were used for the respective average of each class, namely: (1) 20S proteasomes alone (comprising 3.5% of the total number of particles); (2) singly capped 26S proteasomes (comprising 32%); (3) doubly capped 26S particles (31%); (4) the new hybrid proteasomes containing one PA28 ring and one 19S particle at opposite ends of the 20S particle (10%); (5) 20S proteasomes associated with one (2.5%) PA28 ring; or (6) 20S proteasomes associated with two (1.5%) PA28 rings. Approximately 20% of the selected particles could not be classified unambiguously but contained at least one 19S particle and therefore would have belonged to classes 2, 3 or 4. As discussed below, the relatively low amounts of the hybrid particles seen in electron micrographs must underestimate their actual abundance in solution since EM involves relatively harsh procedures (e.g. staining samples) and requires a large dilution of the particles, which should cause the dissociation of PA28α. Consequently, the PAGE pattern (Figure 1A), which indicated quantitative conversion of singly capped proteasomes into hybrid particles, is probably a much better measure of the frequency of hybrid proteasomes than EM.
Fig. 2. (A) Electron micrographs of a negatively stained proteasome mixture showing the six different proteasome species: a 20S proteasome, proteasomes that are singly capped (19S–20S) and doubly capped (19S–20S–19S) with 19S complex, proteasomes that are singly capped (PA28–20S) and doubly capped (PA28–20S–PA28) with PA28 rings as well as a hybrid proteasome with a 19S complex and a PA28 ring bound to either end (19S–20S–PA28). For visualization of the different complexes, an excess of PA28 was used due to the weak interaction of PA28α rings with the 20S proteasome. Accordingly, our images showed many top views of unbound PA28 rings. As a result of the weak PA28α–proteasome interaction, PA28 rings often dissociated from the 20S proteasome upon adsorption to the carbon film and could be seen as a top view adjacent to the proteasome (see, for example, the particle denoted PA28–20S–PA28). Such particles were excluded from the multireference alignment procedure and not used for the calculation of averages. (B) Known forms of the proteasome: gallery of negatively stained particles and the corresponding average images. (1) 20S proteasomes with two 19S caps (19S–20S–19S); (2) proteasomes with one 19S cap (19S–20S); (3) 20S proteasomes (20S); (4) proteasomes with one PA28 cap (20S–PA28); and (5) proteasomes with two PA28 caps (20S–PA28). The corresponding averages contain 106, 748, 164, 78 and 66 particles. The length of the side of the individual frames corresponds to 80 nm. (C) Hybrid proteasome complex (19S–20S–PA28): negatively stained particles and average image based upon 266 particles.
Protein breakdown by hybrid complexes yields distinct products
In order to characterize further the special properties of the 19S–20S–PA28 hybrid complexes, these mixed preparations and ones not containing PA28 were incubated with several denatured proteins of different sizes [insulin-like growth factor-1 (IGF-1), casein and ovalbumin], which we previously have shown are degraded by 26S particles in a linear and ATP-dependent fashion for many hours (Kisselev et al., 1999; Cascio et al., 2001). Interestingly, even though the addition of PA28α (Figure 1A) or PA28αβ (not shown) caused a 3- to 4-fold increase in hydrolysis of several peptides by the 26S particles, the rate of breakdown of the proteins did not change or increased only slightly, as measured by the generation of new peptides by fluorescamine (Figure 3A), a process that is tightly linked to and directly proportional to the disappearance of the polypeptide (Kisselev et al., 1999; Cascio et al., 2001). Under these conditions, the degradation of the proteins was carried out strictly by the 26S or the hybrid complexes since, even after 6 h, these species were the only particles present in our samples on native gels (Figure 3B). This lack of stimulation of protein breakdown upon PA28 addition is consistent with the conclusion that degradation of proteins requires their binding to the 19S particle, which determines the rate of proteolysis.
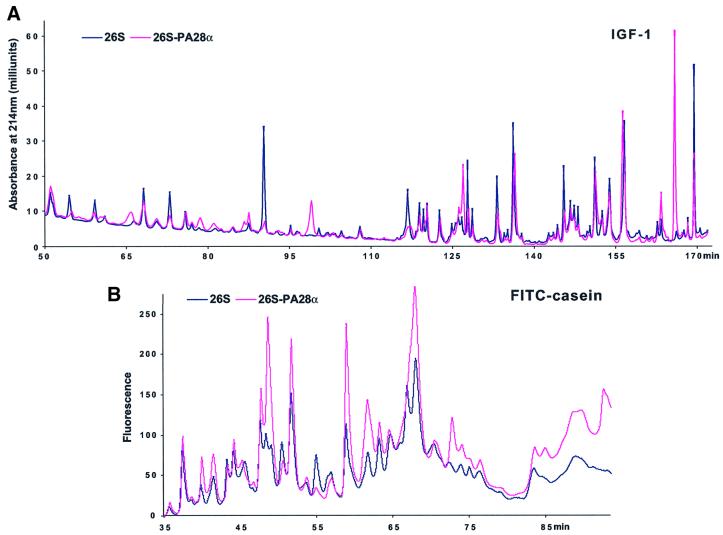
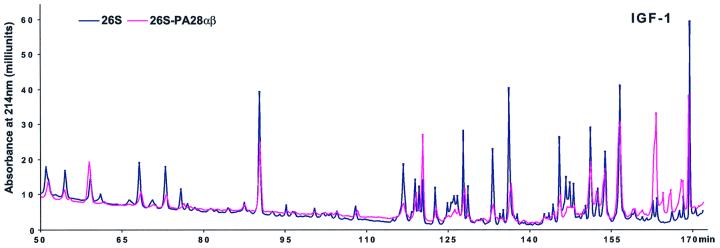
Because the association of PA28 with 19S–20S proteasomes did not alter the rate of protein hydrolysis significantly (as measured by the total amount of new peptides produced), we investigated whether, alternatively, PA28α might modify the pattern of peptides generated from these proteins. Such an effect presumably might account for the proposed ability of PA28 to enhance the generation of peptides suitable for MHC class I presentation. We therefore incubated the 26S and hybrid particles with IGF-1 and fluorescein isothiocyanate (FITC)-labeled casein as described previously (Kisselev et al., 1999) and separated the various peptides produced by reverse-phase HPLC. Under these conditions (e.g. with a large excess of protein substrate), different peptide products are generated at similar linear rates and are not cleaved further again by the 26S proteasomes after their release from the particles (Cascio et al., 2001). The peptides generated by 26S proteasomes from these two substrates clearly differed when PA28α was present and bound to one end of the particle (Figure 4). Moreover, in analogous experiments, addition of native PA28αβ was found to alter the pattern of peptides generated by the 26S particles in a very similar way to PA28α. However, the native heteromeric complex had this effect at a significantly lower concentration; in fact, at the low concentration where PA28αβ was studied (Figure 5), PA28α alone had no effect on the pattern of peptides generated (data not shown). Thus, with either PA28α or PA28αβ added, there were clearly some peptide peaks that were produced only upon formation of the hybrid complexes, and some that were produced in much lower amounts or not at all.
Fig. 4. The association of PA28 with the 26S proteasomes modifies the patterns of peptides generated from proteins. (A) IGF-1 was digested by 26S or hybrid complexes, and peptides generated were separated using a C8 Vydac column as described previously (Cascio et al., 2001), except for the use of an HP 1100 chromatographer (Hewlett-Packard) in the present studies. (B) FITC–casein (10 µM) was degraded as already described (see Figure 3A), and an aliquot was injected onto a C18 Vydac column equilibrated with 10 mM sodium phosphate (pH 6.8) and analyzed by detection of fluorescence. Under these conditions, protein substrate is degraded and peptide products generated at linear rates (Cascio et al., 2001). Peptides were eluted by a gradient of acetonitrile from 0 to 50% in 100 min. Data very similar to those shown in (A) and (B) were obtained in several different analyses.
Fig. 5. Native PA28αβ is able to modify the patterns of peptides generated from IGF-1 in a way similar to recombinant PA28α. Digestion of IGF-1 and separation of peptides were performed as in Figure 4, except that PA28αβ was used at a 3-fold lower concentration than PA28α (0.5 µM instead of 1.5 µM).
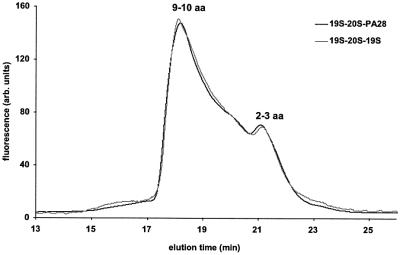
To define further the changes in the pattern of products resulting from binding of PA28 to one end of the 26S particle, the individual peptides generated from IGF-1 were also analyzed by tandem mass spectrometry. By this approach, 34 different peptides were identified unambiguously. They ranged in length from six to 22 residues and were derived from the entire length of this polypeptide (Figure 6). Seventeen of these peptides were generated by both 26S proteasomes and the hybrid complexes (Figure 6A). However, 11 additional peptides were generated only by the 19S–20S–PA28 complexes (Figure 6B).
Fig. 6. Different peptides generated from IGF-1 by 26S proteasomes and hybrid complexes.
In addition, six peptides that were produced by the 26S particles were markedly reduced and could not be detected reliably when PA28 was able to associate with the singly capped 26S proteasomes (Figure 6C). Together, these findings indicate a qualitative change in the cleavages made in proteins upon formation of the hybrid complex.
Product size does not change upon binding of PA28
Recently, Whitby et al. (2000) showed that PA26 binding to the yeast 20S proteasome leads to an opening of the gate that normally seals the pore in the outer (α) rings of the free 20S particles. They therefore suggested that PA28 promotes antigen presentation by allowing premature release of peptides from the proteolytic chamber, which could result in the generation of longer peptides (Kisselev et al., 1999; Kohler et al., 2001). This proposal is attractive since normally most products of the proteasome are shorter than eight residues, which is too small to bind to MHC class I molecules (Kisselev et al., 1999). Also when ovalbumin is degraded by 26S proteasomes, >90% of the time the immunodominant epitope SIINFEKL is cleaved internally (Cascio et al., 2001). Therefore, premature release of products should enhance the yield of longer peptides capable of serving as antigenic precursors. Accordingly, we have found that mutations that leave the gate in the proteasomal α-ring in an open position lead to generation of products of greater mean size (Kohler et al., 2001).
Using the same size-exclusion chromatographic methods as in these prior studies, we tested whether the addition of PA28α might similarly increase the mean size of the products of 26S particles. Surprisingly, despite the clear differences in the patterns of peptides generated from proteins upon formation of the hybrid complexes (Figures 4, 5 and 6), the size distributions of peptides released from 26S proteasomes were indistinguishable in the presence or absence of PA28 (Figure 7). In addition, the mean sizes of peptides produced only by the hybrid complexes that were identified by tandem mass spectrometry (Figure 6) were not significantly different from those generated by the 26S particles lacking PA28.
Fig. 7. Size distribution of peptides generated from IGF-1 by 26S and hybrid complexes. Peptides generated during degradation of IGF-1 were separated from undigested protein on a C18 Vydac column as described (Kisselev et al., 1998), and lyophilized. After lyophilization, the resuspended material was used for size-exclusion chromatography with a polyhydroxy-ethyl aspartamide column (0.46 × 20 cm, Poly LC, Columbia, MD), equilibrated with 0.2 M sodium sulfate, 25% acetonitrile pH 3.0 using an HP1090 chromatographer (Hewlett-Packard) and a fluorometer detector. Peptide products were resuspended in 0.1 M HEPES buffer pH 6.8. For each analysis, 3 nmol of peptides in a total volume of 20 µl were added to 10 µl of fluorescamine (0.3 mg/ml acetone). The reaction was terminated after 1 min with 30 µl of H2O and the sample was injected immediately into the HPLC column. To determine the apparent molecular mass of the peptides eluted, the column was calibrated each time before use with 11 standard amino acids and peptides in the 200–3500 Da range that had been derivatized with fluorescamine in the same way as proteasome products. Prior control studies showed that retention times of these fluorescamine-derivatized peptides were highly reproducible, and linearly dependent on the logarithm of their molecular weights (Kohler et al., 2001).
Discussion
Although immunoprecipitation studies had clearly indicated the existence in vivo of a 19S–20S–PA28 complex (Hendil et al., 1998) that is induced by γ-interferon (Tanahashi et al., 2000), prior attempts to isolate this hybrid particle from cells or tissues have failed repeatedly, apparently because of its lability and sensitivity to salt (Tanahashi et al., 2000). As an alternative approach to define its structure and properties, we have established conditions that allow its in vitro reconstitution (see also Kopp et al., 2001). To quantitatively convert the singly capped particles into hybrid complexes (Figure 1), a large molar excess (30:1) of PA28α over 26S proteasomes was required. Thus, the affinity of the singly capped 26S for PA28α, like that of 20S particles, is not high, and the complex dissociates readily upon dilution (see below). In vivo, additional factors probably facilitate the association between the core particle and PA28, and a role for phosphorylation in this process has been suggested (Dubiel et al., 1992; Yang et al., 1995).
Although PA28β alone, at the same concentration as PA28α, does not stimulate peptide hydrolysis (P.Cascio, unpublished observations), the presence of β-subunits in the natural complex PA28αβ can increase the affinity of PA28 rings for the 20S proteasome (Song et al., 1997). Accordingly, in the present studies, the Kd for the activation of the 26S particles by the natural PA28αβ was 5-fold lower than with PA28α. In addition, the heteromeric complex caused alterations very similar to those caused by PA28α in the pattern of peptides generated from the protein substrates, but the natural molecule did so at significantly lower concentrations (see below and Figure 5). It is noteworthy that the Kd for the association of PA28α with the 26S particles and the resulting activation of peptide hydrolysis were indistinguishable with 26S proteasomes and immunoproteasomes. Therefore, the presence of the γ-interferon-induced β-subunits in place of the constitutive ones has no effect on the affinity of the core particle for PA28.
It is also noteworthy that after incubation with a large excess of PA28α, the band of singly capped 26S proteasomes disappeared on native gels and formed larger structures, without any detectable generation of 20S particles containing two PA28 complexes. Therefore, PA28 is not able to bind to 20S as strongly as the 19S complex (Hoffman and Rechsteiner, 1994; Yang et al., 1995), whose association with the 20S requires ATP (Chu-Ping et al., 1994; DeMartino et al., 1994) and even withstands high concentrations of salt (Kania et al., 1996). Instead, PA28 seems to associate with the free end of an asymmetric 19S–20S complex, as also indicated by EM. No such hybrid complex was seen in the absence of PA28. Although native PAGE indicated that all the singly capped structures (approximately half the 26S proteasomes) were converted to hybrid complexes, by EM these hybrid structures appeared to comprise 10% (or more) of the different forms on the grid. However, this latter value certainly underestimates the actual abundance of hybrid particles in solution, because the large dilution of samples necessary to obtain clear EM images causes dissociation of PA28 (especially of PA28α). In addition, the procedures for the absorption of particles to the grids and negative staining of the sample probably result in further dissociation of PA28 (and even of some 19S) from the 20S particles.
Also, if only 10% of the 26S particles formed PA28–20S–19S complexes, it is quite unlikely that a 3- to 4-fold increase in total peptide hydrolysis upon addition of PA28 could result. To explain such a stimulation, it is instead much more likely that PA28α caused quantitative formation of hybrid complexes from the 50% of the proteasomes that were singly capped. Accordingly, the native PAGE (Figure 1B) and the activity assays on the gels indicate that nearly all of the singly capped particles were in fact converted into more active larger complexes by PA28. Together, these findings argue further that most of the hybrid particles present in solution were lost during the EM analysis.
Protein breakdown by hybrid complexes
After PA28 associated with the free end of the 19S–20S complex, the resulting particle, though more active against fluorogenic peptides, did not show a significantly greater capacity for protein breakdown. Therefore, the rate of protein hydrolysis must be determined by the 19S complex, and proteolysis by the hybrid complex appears to be a vectoral process, in which protein substrates are bound initially by the 19S component, where they are unfolded (Braun et al., 1999; Benaroudj and Goldberg, 2000; Navon and Goldberg, 2001) and translocated into the proteolytic chamber for hydrolysis. Then the peptide products are released mainly from the other end of the proteasome through the PA28 ring. This asymmetric 19S–20S organization can obviously explain how PA28 binding to the free end of the 20S barrel causes a selective increase in the hydrolysis of small peptides, which, unlike proteins, should enter the particle easily through the PA28-induced open channel.
In the vectoral degradation of proteins, PA28, by simply promoting opening of the gate that normally blocks the exit of peptides from the proteolytic chamber, would be expected to cause premature release of some longer peptides (Kohler et al., 2001). Most products released from the proteasomes are too small to serve in antigen presentation, i.e. <8–9 residues (Kisselev et al., 1999), and premature release of incompletely digested fragments should, in principle, increase the yield of longer peptides capable of binding directly to MHC class I molecules or being trimmed to the appropriate length by aminopeptidases (Cascio et al., 2001). In fact, we recently demonstrated a clear increase in the sizes of peptides generated by mutant yeast proteasomes with a constitutively open gate in the α-ring (Kohler et al., 2001). However, in the present studies, using the same analytical approach (and also by use of mass spectrometry), we were not able to find any change in the size distribution of 26S proteasome products in the presence of PA28. This failure of PA28 to increase product size implies that it causes gate opening to a lesser extent than the yeast α-deletion mutation.
This lack of any clear change in product size, despite the qualitative changes in the nature of the peptide products, may appear surprising. However, it is noteworthy that the 19S complexes in the doubly capped particles also open the gate in the 20S particle (Kohler et al., 2001), and there is no reason to assume that its opening by PA28 would result in a different size distribution of the peptides’ products than would be achieved upon association of 20S proteasomes with two 19S particles. It is also possible that because the preparations studied here contained a mixed population of hybrid complexes and doubly capped 26S particles, we may have been unable to detect a small increase in the sizes of peptide products. However, in vivo, hybrid complexes are also not present as pure populations and, even after prolonged stimulation with γ-interferon, they appear to represent only 25% of the total content of proteasomes in HeLa cells (Tanahashi et al., 2000). Thus, a clear change in mean product size seems unlikely in vivo.
An attractive alternative explanation of these results is that PA28, in addition to influencing gate opening, can allosterically modify the active sites of the core particle in a manner that increases the generation of peptides suitable for MHC class I presentation. Although no modification in the structure of the yeast 20S active sites was found upon association with the trypanosome PA26 (Whitby et al., 2000), these studies were at low resolution, and this interaction may also not mimic exactly the changes induced by mammalian PA28 on mammalian 20S proteasomes. In fact, recent observations by Li and Rechsteiner (2001) indicate that binding of mammalian PA28γ to 20S particles results in a selective allosteric stimulation of only the trypsin-like activity, which argues strongly against the conclusion that these ring complexes only facilitate opening of the gate (which would result in an increased activity of all three catalytic sites). Thus, PA28 appears both to enhance certain cleavages and facilitate the release of peptide products, so that although the mean product size does not change, a different and presumably broader range of peptides is generated, as we found here.
In fact, while the size distribution of the products of the 26S proteasome was not affected by formation of the hybrid 19S–20S–PA28 complex (Figure 7), the patterns of peptides generated from the two protein substrates changed dramatically, as shown by reverse-phase HPLC with either PA28α (Figure 4) or native PA28αβ (see Figure 5 for details). These changes in peptide production were confirmed by mass spectrometry sequencing, which showed that while many peptides were generated both by 26S and hybrid proteasomes (Figure 6A), several were generated only if PA28α was also present (Figure 6B). Specifically, only the 19S–20S–PA28 complexes were found to generate certain peptides from the N-terminal part of IGF-1, perhaps by preventing the complete degradation of this region to di/tripeptides. This release of new products thus seems to result from PA28α-induced alterations in how the 26S proteasome cleaves polypeptide chains.
Several observations argue that this marked change in the pattern of products is not simply due to the 19S–20S–PA28 complexes taking up and degrading further certain peptides previously released by the 26S particles. Such a mechanism would require selective uptake and hydrolysis of certain very dilute peptides, and would result in an overall decrease in the mean size of peptide products after binding of PA28, which was not observed (Figure 7). Also, our extensive prior control studies had indicated that under these incubation conditions, the production of different peptides occurred at linear, parallel rates with no further metabolism of these products after their release from the 26S particles (Cascio et al., 2001). Finally, of the 11 new peptides generated by the 19S–20S–PA28α complexes from IGF-1, nearly half were not even present in the various peptide products of the 26S particles (Figure 6A and B). Their generation must have involved a different primary proteolytic attack of IGF-1. On the other hand, six of the products of IGF-1 degradation by 26S proteasomes could not be detected when PA28 was also present (Figure 6C). Taken together, these data argue strongly that the binding of PA28 directly modifies the cleavage properties of the 26S proteasomes, so that a different spectrum of peptides is generated. In this way, PA28 would enhance the variability of epitopes presented on the cell surface and, thereby, increase the possibility that an appropriate CTL response is elicited.
Materials and methods
Protein purification
20S and 26S immunoproteasomes were purified from rabbit spleen as described previously and are free of aminopeptidases that may act on proteasome products (Cascio et al., 2001). Recombinant mouse PA28α was expressed in E.coli, and purified as described (Song et al., 1997) with minor modifications. Briefly, after isolation of the inclusion bodies, the precipitated protein was denaturated and solubilized in buffer containing 8 M urea and 0.1 M dithiothreitol (DTT). The urea was then removed by gel filtration and at this stage the protein was mostly soluble and able to stimulate by several fold the peptidase activities of the 20S proteasome. Purification to apparent homogeneity was achieved using a Mono-Q HR column (Amersham Pharmacia) followed by gel filtration on a Superose 6 HR column (Amersham Pharmacia). Native rabbit PA28αβ was purchased from Boston Biochem (Cambridge, MA) or Affinity (Mamhead, UK).
Peptidase assay and electrophoretic methods
Continuous measurements of proteasome activity were performed as described previously (Kisselev et al., 1999). Native PAGE of proteasomes was performed according to established techniques, except that the gel and running buffers contained 25 mM Tris–borate pH 8.3, 0.5 mM EDTA, 1.25 mM MgCl2 and 0.25 mM ATP.
Reconstitution of the hybrid complex
To reconstitute the hybrid complex, the 26S proteasomes (50 nM) were pre-incubated with PA28α (1.5 µM) at 37°C for 30 min in 20 mM Tris–HCl pH 7.5, 2 mM ATP, 5 mM MgCl2, 1 mM EDTA. Such samples were then analyzed directly by native PAGE or used for EM or studies of protein degradation. In studies of products of hybrid complexes containing native PA28αβ, this protein was used at a 3-fold lower concentration than PA28α (see Figure 5).
Electron microscopy
Samples of the hybrid complex were diluted 1:15 with incubation buffer immediately prior to adsorption onto glow-discharged carbon-coated copper grids. Grids were washed with four drops of incubation buffer and stained with two drops of freshly prepared 0.75% uranyl formate (Pfaltz & Bauer, Waterbury, CT). Specimens were inspected with a Philips Tecnai 12 electron microscope operated at 120 kV and images were taken at a nominal magnification of 52 000× using low-dose procedures. A total of 133 images were digitized with a Zeiss SCAI scanner using a pixel size of 6.7 Å at the specimen level, from which 4595 particles were selected for further computational processing using the SPIDER image processing package (Frank et al., 1996). In a first step, particles were grouped into six classes corresponding to free 20S proteasomes (164 particles) and 20S particles singly capped with one 19S (1467 particles), doubly capped with two 19S (1419 particles), singly capped with one PA28 ring (123 particles), doubly capped with two PA28 rings (66 particles) and proteasomes associated with one 19S complex and one PA28 ring (491 particles). In a second step, each class was subjected independently to a multireference alignment procedure and, from the resulting class averages, the average with the sharpest features was chosen.
Protein digestions and peptide analysis
Methods for oxidation of ovalbumin (Kisselev et al., 1999) and carboxymethylation of IGF-1 (Akopian et al., 1997) to ensure denaturation and to prevent sulfhydryl bond formation were described previously. Ovalbumin, IGF-1 and casein were then reductively methylated to reduce the background in the fluorescamine assay (Akopian et al., 1997), and digestion by proteasomes was performed as described previously (Cascio et al., 2001). Under these incubation conditions (i.e. with a large excess of protein substrate), proteins are degraded and different peptide products are generated by 26S proteasomes at similar linear rates for many hours (Cascio et al., 2001). As demonstrated previously, the peptides, once generated, do not undergo significant further digestion by the proteasomes, and the relative amounts of different products did not change.
To assay peptides generated during protein degradation, we measured the appearance of new amino groups using fluorescamine. FITC–casein was prepared (Akopian et al., 1997) and its degradation by proteasomes measured at different times. At the end of the incubation, the undigested FITC–casein was then precipitated with 2% perchloric acid, and the fluorescence in the supernatant was measured (λ excitation 490 nm, λ emission 525 nm). Reverse-phase HPLC analysis of peptides generated by proteasomes and hybrid complexes from IGF-1 and FITC–casein were performed using methods described elsewhere (Akopian et al., 1997; Kisselev et al., 1999). Size-exclusion chromatography of peptides generated from IGF-1 was performed using a new method (Kohler et al., 2001) in which the total pool of peptides produced was derivatized with fluorescamine before injection in the HPLC column and the fluorescence that was eluted from the column was monitored continuously (see Figure 7 for more details).
Peptide analysis by mass spectrometry
Peptides generated by the 26S proteasomes and the hybrid complexes were subjected to sequencing by tandem mass spectrometry on a Thermo Finningan LCQ Deca Quadrupole Ion Trap Mass Spectrometer.
Acknowledgments
Acknowledgements
We thank S.Gygi for his expert assistance in peptide analysis by mass spectrometry, A.Navon for help in the preparation of PA28α, and J.Frisbie for valuable assistance in preparing this manuscript. The molecular electron microscopy facility at Harvard Medical School was established by a generous gift from the Giovanni Armenise Harvard Center for Structural Biology and is maintained by funds from NIH grant P01 GM62580-01. These studies were supported by grants from the NIGMS to A.L.G. P.C. also received support from the Giovanni Armenise Foundation.
References
- Adams G.M., Falke,S., Goldberg,A.L., Slaughter,C.A., DeMartino,G.N. and Gogol,E.P. (1997) Structural and functional effects of PA700 and modulator protein on proteasomes. J. Mol. Biol., 273, 646–657. [DOI] [PubMed] [Google Scholar]
- Ahn J.Y. et al. (1995) Primary structures of two homologous subunits of PA28, a γ-interferon-inducible protein activator of the 20S proteasome. FEBS Lett., 366, 37–42. [DOI] [PubMed] [Google Scholar]
- Ahn K., Erlander,M., Leturcq,D., Peterson,P.A., Fruh,K. and Yang,Y. (1996) In vivo characterization of the proteasome regulator PA28. J. Biol. Chem., 271, 18237–18242. [DOI] [PubMed] [Google Scholar]
- Akopian T.N., Kisselev,A.F. and Goldberg,A.L. (1997) Processive degradation of proteins and other catalytic properties of the proteasome from Thermoplasma acidophilum. J. Biol. Chem., 272, 1791–1798. [DOI] [PubMed] [Google Scholar]
- Benaroudj N. and Goldberg,A.L. (2000) PAN, the proteasome-activating nucleotidase from archaebacteria, is a protein-unfolding molecular chaperone. Nature Cell Biol., 2, 833–839. [DOI] [PubMed] [Google Scholar]
- Beninga J., Rock,K.L. and Goldberg,A.L. (1998) Interferon-γ can stimulate post-proteasomal trimming of the N terminus of an antigenic peptide by inducing leucine aminopeptidase. J. Biol. Chem., 273, 18734–18742. [DOI] [PubMed] [Google Scholar]
- Braun B.C., Glickman,M., Kraft,R., Dahlmann,B., Kloetzel,P.M., Finley,D. and Schmidt,M. (1999) The base of the proteasome regulatory particle exhibits chaperone-like activity. Nature Cell Biol., 1, 221–226. [DOI] [PubMed] [Google Scholar]
- Cascio P., Hilton,C., Kisselev,A.F., Rock,K.L. and Goldberg,A.L. (2001) 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. EMBO J., 20, 2357–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu-Ping M., Vu,J.H., Proske,R.J., Slaughter,C.A. and DeMartino,G.N. (1994) Identification, purification and characterization of a high molecular weight, ATP-dependent activator (PA700) of the 20S proteasome. J. Biol. Chem., 269, 3539–347. [PubMed] [Google Scholar]
- Craiu A., Akopian,T., Goldberg,A. and Rock,K.L. (1997) Two distinct proteolytic processes in the generation of a major histocompatibility complex class I-presented peptide. Proc. Natl Acad. Sci. USA, 94, 10850–10855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMartino G.N., Moomaw,C.R., Zagnitko,O.P., Proske,R.J., Chu-Ping, M., Afendis,S.J., Swaffield,J.C. and Slaughter,C.A. (1994) PA700, an ATP-dependent activator of the 20S proteasome, is an ATPase containing multiple members of a nucleotide-binding protein family. J. Biol. Chem., 269, 20878–20884. [PubMed] [Google Scholar]
- Dick T.P., Ruppert,T., Groettrup,M., Kloetzel,P.M., Kuehn,L., Koszinowski,U.H., Stevanovic,S., Schild,H. and Rammensee,H.G. (1996) Coordinated dual cleavages induced by the proteasome regulator PA28 lead to dominant MHC ligands. Cell, 86, 253–262. [DOI] [PubMed] [Google Scholar]
- Dubiel W., Pratt,G., Ferrell,K. and Rechsteiner,M. (1992) Purification of an 11S regulator of the multicatalytic protease. J. Biol. Chem., 267, 22369–22377. [PubMed] [Google Scholar]
- Eleuteri A.M., Kohanski,R.A., Cardozo,C. and Orlowski,M. (1997) Bovine spleen multicatalytic proteinase complex (proteasome). Replacement of X, Y and Z subunits by LMP7, LMP2 and MECL1 and changes in properties and specificity. J. Biol. Chem., 272, 11824–11831. [DOI] [PubMed] [Google Scholar]
- Frank J., Radermacher,M., Penczek,P., Zhu,J., Li,Y., Ladjadj,M. and Leith,A. (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol., 116, 190–199. [DOI] [PubMed] [Google Scholar]
- Groettrup M., Soza,A., Eggers,M., Kuehn,L., Dick,T.P., Schild,H., Rammensee,H.G., Koszinowski,U.H. and Kloetzel,P.M. (1996) A role for the proteasome regulator PA28α in antigen presentation. Nature, 381, 166–168. [DOI] [PubMed] [Google Scholar]
- Groll M., Bajorek,M., Kohler,A., Moroder,L., Rubin,D.M., Huber,R., Glickman,M.H. and Finley,D. (2000) A gated channel into the proteasome core particle. Nature Struct. Biol., 7, 1062–1067. [DOI] [PubMed] [Google Scholar]
- Hendil K.B., Khan,S. and Tanaka,K. (1998) Simultaneous binding of PA28 and PA700 activators to 20S proteasomes. Biochem. J., 332, 749–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman L. and Rechsteiner,M. (1994) Activation of the multicatalytic protease. The 11S regulator and 20S ATPase complexes contain distinct 30-kilodalton subunits. J. Biol. Chem., 269, 16890–16895. [PubMed] [Google Scholar]
- Kania M.A., Demartino,G.N., Baumeister,W. and Goldberg,A.L. (1996) The proteasome subunit, C2, contains an important site for binding of the PA28 (11S) activator. Eur. J. Biochem., 236, 510–516. [DOI] [PubMed] [Google Scholar]
- Kisselev A.F., Akopian,T.N. and Goldberg,A.L. (1998) Range of sizes of peptide products generated during degradation of different proteins by archaeal proteasomes. J. Biol. Chem., 273, 1982–1989. [DOI] [PubMed] [Google Scholar]
- Kisselev A.F., Akopian,T.N., Woo,K.M. and Goldberg,A.L. (1999) The sizes of peptides generated from protein by mammalian 26 and 20S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J. Biol. Chem., 274, 3363–3371. [DOI] [PubMed] [Google Scholar]
- Knowlton J.R., Johnston,S.C., Whitby,F.G., Realini,C., Zhang,Z., Rechsteiner,M. and Hill,C.P. (1997) Structure of the proteasome activator REGα (PA28α). Nature, 390, 639–643. [DOI] [PubMed] [Google Scholar]
- Kohler A., Cascio,P., Leggett,D.S., Woo,K.M., Goldberg,A.L. and Finley,D. (2001) The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol. Cell, 7, 1143–1152. [DOI] [PubMed] [Google Scholar]
- Kopp F., Dahlmann,B. and Kuehn,L. (2001) Reconstitution of hybrid proteasomes from purified PA700–20S complexes and PA28αβ activator: ultrastructure and peptidase activities. J. Mol. Biol., 313, 465–471. [DOI] [PubMed] [Google Scholar]
- Kuehn L. and Dahlmann,B. (1996) Proteasome activator PA28 and its interaction with 20S proteasomes. Arch. Biochem. Biophys., 329, 87–96. [DOI] [PubMed] [Google Scholar]
- Li J. and Rechsteiner,M. (2001) Molecular dissection of the 11S REG (PA28) proteasome activators. Biochimie, 83, 373–383. [DOI] [PubMed] [Google Scholar]
- Ma C.P., Slaughter,C.A. and DeMartino,G.N. (1992) Identification, purification and characterization of a protein activator (PA28) of the 20S proteasome (macropain). J. Biol. Chem., 267, 10515–10523. [PubMed] [Google Scholar]
- Mo A.X., van Lelyveld,S.F., Craiu,A. and Rock,K.L. (2000) Sequences that flank subdominant and cryptic epitopes influence the proteolytic generation of MHC class I-presented peptides. J. Immunol., 164, 4003–4010. [DOI] [PubMed] [Google Scholar]
- Murata S. et al. (2001) Immunoproteasome assembly and antigen presentation in mice lacking both PA28α and PA28β. EMBO J., 20, 5898–5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navon A. and Goldberg,A.L. (2001) Proteins are unfolded on the surface of the ATPase ring before transport into the proteasome. Mol. Cell, 8, 1339–1349. [DOI] [PubMed] [Google Scholar]
- Preckel T. et al. (1999) Impaired immunoproteasome assembly and immune responses in PA28–/– mice. Science, 286, 2162–2165. [DOI] [PubMed] [Google Scholar]
- Realini C., Dubiel,W., Pratt,G., Ferrell,K. and Rechsteiner,M. (1994) Molecular cloning and expression of a γ-interferon-inducible activator of the multicatalytic protease. J. Biol. Chem., 269, 20727–20732. [PubMed] [Google Scholar]
- Rechsteiner M., Realini,C. and Ustrell,V. (2000) The proteasome activator 11S REG (PA28) and class I antigen presentation. Biochem. J., 345, 1–15. [PMC free article] [PubMed] [Google Scholar]
- Rock K.L. and Goldberg,A.L. (1999) Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu. Rev. Immunol., 17, 739–779. [DOI] [PubMed] [Google Scholar]
- Rock K.L., Gramm,C., Rothstein,L., Clark,K., Stein,R., Dick,L., Hwang,D. and Goldberg,A.L. (1994) Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell, 78, 761–771. [DOI] [PubMed] [Google Scholar]
- Saric T., Beninga,J., Graef,C.I., Akopian,T.N., Rock,K.L. and Goldberg,A.L. (2001) Major histocompatibility complex class I-presented antigenic peptides are degraded in cytosolic extracts primarily by thimet oligopeptidase. J. Biol. Chem., 276, 36474–36481. [DOI] [PubMed] [Google Scholar]
- Schwarz K., van Den Broek,M., Kostka,S., Kraft,R., Soza,A., Schmidtke,G., Kloetzel,P.M. and Groettrup,M. (2000) Overexpression of the proteasome subunits LMP2, LMP7 and MECL-1, but not PA28α/β, enhances the presentation of an immunodominant lymphocytic choriomeningitis virus T cell epitope. J. Immunol., 165, 768–778. [DOI] [PubMed] [Google Scholar]
- Shimbara N., Nakajima,H., Tanahashi,N., Ogawa,K., Niwa,S., Uenaka,A., Nakayama,E. and Tanaka,K. (1997) Double-cleavage production of the CTL epitope by proteasomes and PA28: role of the flanking region. Genes Cells, 2, 785–800. [DOI] [PubMed] [Google Scholar]
- Snyder H.L., Yewdell,J.W. and Bennink,J.R. (1994) Trimming of antigenic peptides in an early secretory compartment. J. Exp. Med., 180, 2389–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Mott,J.D., von Kampen,J., Pramanik,B., Tanaka,K., Slaughter,C.A. and DeMartino,G.N. (1996) A model for the quaternary structure of the proteasome activator PA28. J. Biol. Chem., 271, 26410–26417. [DOI] [PubMed] [Google Scholar]
- Song X., von Kampen,J., Slaughter,C.A. and DeMartino,G.N. (1997) Relative functions of the α and β subunits of the proteasome activator, PA28. J. Biol. Chem., 272, 27994–28000. [DOI] [PubMed] [Google Scholar]
- Stoltze L. et al. (2000) Two new proteases in the MHC class I processing pathway. Nature Immunol., 1, 413–418. [DOI] [PubMed] [Google Scholar]
- Tanahashi N., Murakami,Y., Minami,Y., Shimbara,N., Hendil,K.B. and Tanaka,K. (2000) Hybrid proteasomes. Induction by interferon-γ and contribution to ATP-dependent proteolysis. J. Biol. Chem., 275, 14336–14345. [DOI] [PubMed] [Google Scholar]
- Thrower J.S., Hoffman,L., Rechsteiner,M. and Pickart,C.M. (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J., 19, 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanKaer L., Ashton-Rickardt,P.G., Eichelberger,M., Gaczynska,M., Nagashima,K., Rock,K.L., Goldberg,A.L., Doherty,P.C. and Tonegawa,S. (1994) Altered peptidase and viral-specific T cell response in LMP2 mutant mice. Immunity, 1, 533–5341. [DOI] [PubMed] [Google Scholar]
- Voges D., Zwickl,P. and Baumeister,W. (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem., 68, 1015–1068. [DOI] [PubMed] [Google Scholar]
- Walz J., Erdmann,A., Kania,M., Typke,D., Koster,A.J. and Baumeister,W. (1998) 26S proteasome structure revealed by three-dimensional electron microscopy. J. Struct. Biol., 121, 19–29. [DOI] [PubMed] [Google Scholar]
- Whitby F.G., Masters,E.I., Kramer,L., Knowlton,J.R., Yao,Y., Wang,C.C. and Hill,C.P. (2000) Structural basis for the activation of 20S proteasomes by 11S regulators. Nature, 408, 115–120. [DOI] [PubMed] [Google Scholar]
- Yang Y., Fruh,K., Ahn,K. and Peterson,P.A. (1995) In vivo assembly of the proteasomal complexes, implications for antigen processing. J. Biol. Chem., 270, 27687–27694. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Krutchinsky,A., Endicott,S., Realini,C., Rechsteiner,M. and Standing,K.G. (1999) Proteasome activator 11S REG or PA28: recombinant REGα/REGβ hetero-oligomers are heptamers. Biochemistry, 38, 5651–5658. [DOI] [PubMed] [Google Scholar]