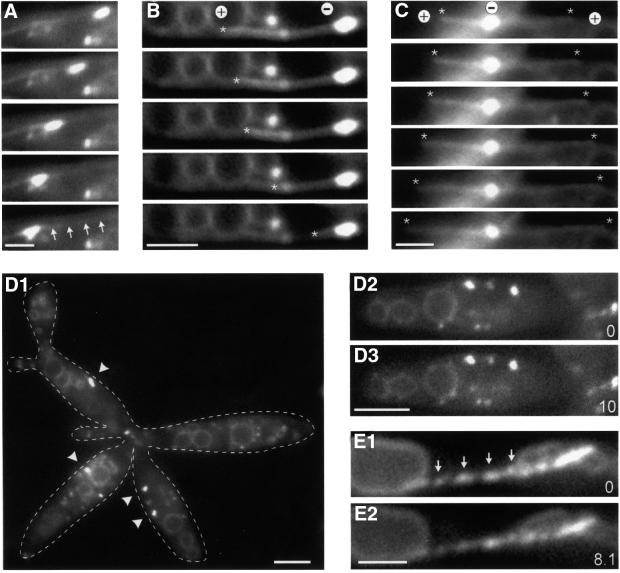
Fig. 5. Motion of EEs in the kin3 deletion strain RWS13. (A–C) Co-expression of a GFP–Tub1 fusion protein and Yup1–GFP in a Δkin3 strain allowed us to analyze the role of MTs in the remaining motility of EEs. Brightly stained EE clusters move along MTs (A; arrows mark the MT). In the absence of Kin3, they were located at the ends of MTs. In all cases (n = 31), MTs rapidly shortened towards these cluster (B; end of MT marked by asterisk), suggesting that EEs are located at the minus-ends (labeled ‘+’ and ‘–’). Consistent with this, MT elongation was found to be directed away from the BSDs (C; ends of MTs marked by asterisks, orientation indicated by ‘+’ and ‘–’). Time between frames is 1.4 s. Bar: 2 µm. (D) Endosome organization in the temperature-sensitive dynein–Δkin3 double mutant (RWS14). After 2 h at 33°C, endosome clusters (arrowheads) are smaller or even absent from the cell (D1; right cell). Note that cells have a separation defect due to the deletion of kin3. In these structures, almost no EE motility was found (D2, D3; time in seconds is given in the bottom right corner). Bars: 3 µm. (E) Expression of a dominant-negative kin3 mutant allele that is described to result in a rigid binding of the motor to the MT leads to ‘pearl-string’-like arrangement of EEs (arrows) and abolished almost all motion (compare E1 and E2). Bar: 2 µm; time in seconds is given in the bottom right corner. Movies are available as Supplementary data.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.