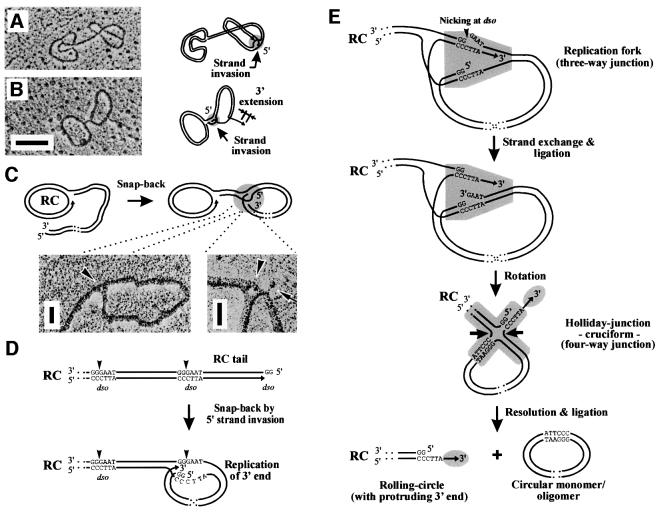
Fig. 8. Model for the resolution of mp1 concatemers. (A and B) Two representative examples of mp1 in vivo circles interconnected by a double-stranded tail. Hypothetical schemes depict a snap-back mechanism using invasion of the ends of the RC tails (grey circles) to release mp1 monomer units (1.3 kb). Bar = 0.5 kb. (C) Hypothetical model and enlarged sections of snap-back junctions (arrowheads) that were recently regarded as strand-switching intermediates (Backert, 2000). The arrow shows a free ssDNA end originating from the junction. Bar = 100 bases. (D) Mp1 concatemers produced by RCR are head-to-tail molecules with specific ends (dso1 or dso2). The 5′ overhang of the RC tail may invade the next dso to form a snap-back molecule. (E) After invasion, a replication fork (shaded grey) is formed in which the 3′ end extends. Resolution of this intermediate proceeds by nicking of the dso motif and strand exchange. This results in a Holliday structure that can easily be resolved by the recombination machinery (arrows). Finally, circular mp1 mono- or oligomers are released from the replicating RC that retains a protruding 3′ end (grey circle).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.