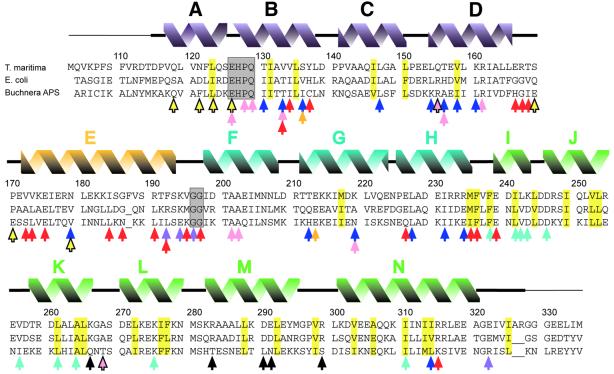
Fig. 2. Alignment of FliG-MC sequences from T.maritima, E.coli and Buchnera APS. The initiating Met in the T.maritima protein studied here replaces a Leu in the native protein. Secondary structural elements seen in T.maritima FliG-MC are shown above. A thin line denotes disordered segments not included in the refined model. Residues that form the cores of domains I and II are highlighted in yellow. Conserved EHPQ and GG motifs discussed in the text are in gray boxes. Black arrows indicate charged residues important for motor rotation in E.coli (Lloyd and Blair, 1997). Colored arrows indicate positions of mutations giving various phenotypes. Turquoise, nonmotile but flagellate (Irikura et al., 1993; Lloyd et al., 1996). Pink, nonflagellate (in E.coli) (Lloyd et al., 1996; Zhou et al., 1998b). Blue, weakened binding to FliM in a yeast two-hybrid assay (Marykwas and Berg, 1996). Red, CW motor bias (Irikura et al., 1993; Lloyd and Blair, 1997). Yellow with black outline, CCW motor bias (Irikura et al., 1993). Orange, either CW or CCW motor bias depending on the substitution (Irikura et al., 1993). Purple, suppressors of mutations in the stator protein MotB (Garza et al., 1996). Most of the point mutations that gave nonflagellate phenotype in E.coli (pink arrows) gave an immotile but flagellate phenotype in Salmonella (Irikura et al., 1993); two exceptions that gave nonflagellate phenotype in both species are indicated by pink arrows with black outline. Sequences are from DDBJ/EMBL/GenBank.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.