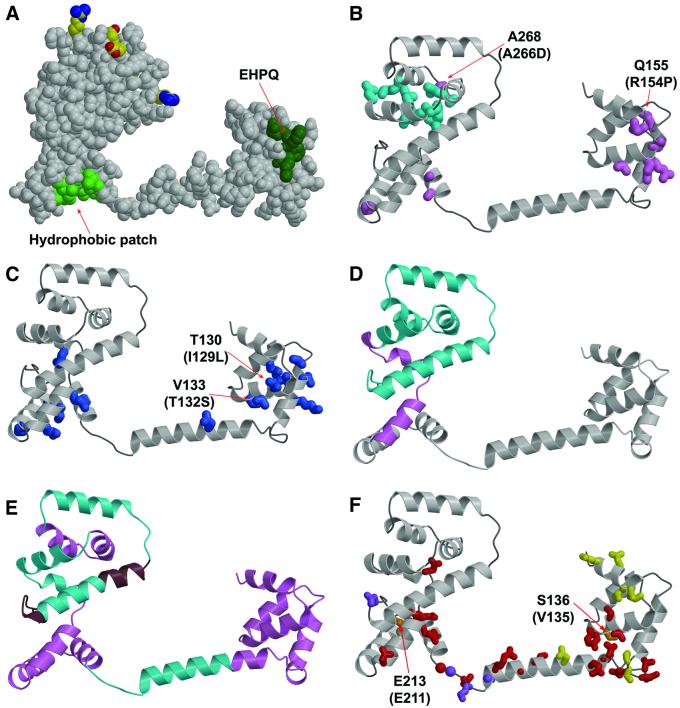
Fig. 5. Mapping of mutations and deletions to the structure. (A) Conserved surface features. Dark green, EHPQ motif in domain I. Light green, surface hydrophobic patch on domain II. Charged residues that interact with the stator are shown in standard atom colors. (B) Turquoise, mutations that eliminate motor rotation but allow flagellar assembly. Pink, mutations that disrupt flagellar assembly in E.coli. Mutations indicated by the labels (Q155 and A268) also disrupt flagellar assembly in Salmonella (compare with the legend to Figure 2; the mutational changes in Salmonella are given in parentheses). (C) Blue, mutations that weaken binding to FliM in the yeast two-hybrid assay (Marykwas and Berg, 1996). Two mutations that involved relatively minor changes in surface-exposed side-chains are indicated, with the mutational changes in the E.coli protein given in parentheses. These two mutations are adjacent to the EHPQ motif. (D) Turquoise, an 86-residue C-terminal segment that can be deleted while still allowing flagellar assembly and binding of FliG to FliM and FliN. Pink, a 19-residue segment whose further deletion prevents flagellar assembly and weakens binding to FliM and FliN (Tang et al., 1996). (E) Turquoise, segments of the protein in which 10-residue deletions prevent motor rotation but allow flagellar assembly. Pink, segments of the protein in which 10-residue deletions prevent flagellar assembly. Brown, segments of the protein in which 10-residue deletions allow both assembly and rotation. (F) Red, mutations giving CW motor bias (Irikura et al., 1993; Lloyd and Blair, 1997). Yellow, mutations giving CCW motor bias (Irikura et al., 1993). Orange, two positions that, depending on the substitution, give either CW or CCW motor bias; the residue numbers for T.maritima and Salmonella are shown (Salmonella in parentheses) (Irikura et al., 1993). Purple, mutations that suppress motility defects caused by mutations in the stator protein MotB (Garza et al., 1996).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.