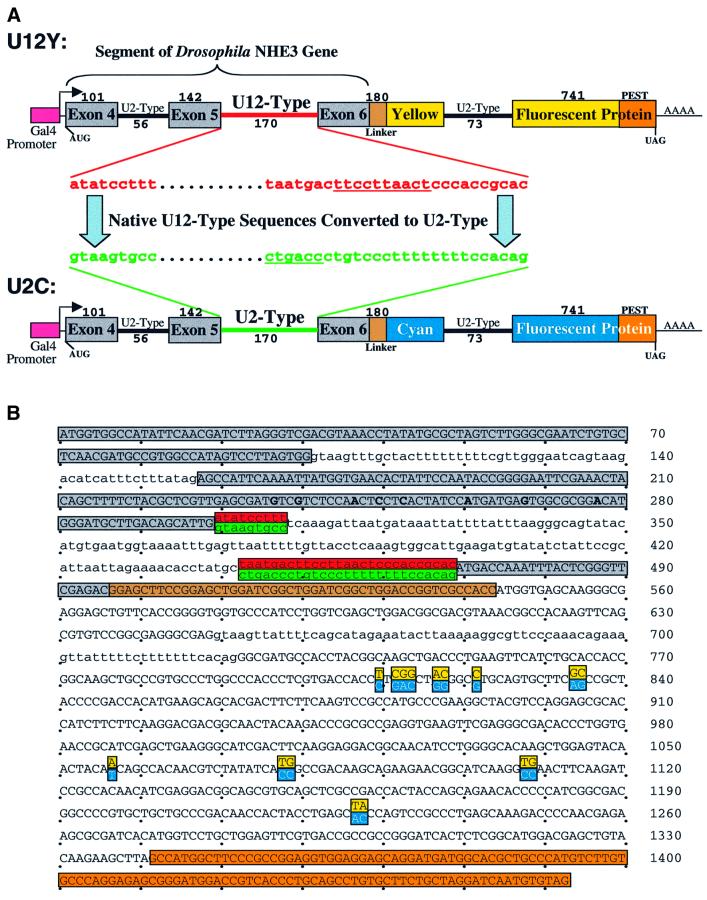
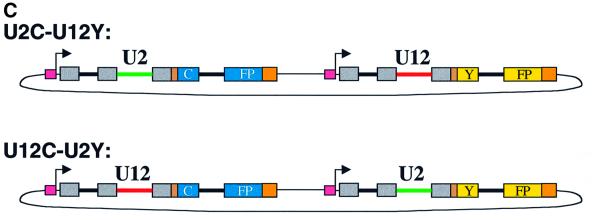
Fig. 3. Plasmids that report the efficiency of U12-type versus U2-type splicing. (A) A segment of the D.melanogaster NHE3 gene including introns 4 (U2-type) and 5 (U12-type) and their flanking exons was fused, via a linker, to a yellow (Y) or cyan (C) fluorescent protein coding sequence containing destabilizing ‘PEST’ sequences to form U12Y or U12C. Intron 6 (U2-type) of NHE3 was inserted into the CFP or YFP coding sequences. A Gal4-responsive promoter and a Kozak consensus translation start site were directly upstream of the fusion protein. Exon and intron lengths are indicated above and below (not drawn to scale). To form U2Y (or U2C), consensus splicing sequences of the U12-type intron were mutated to U2-type sequences derived from an adenovirus intron, keeping the intron length constant. (B) The sequences of the protein-coding regions of the fusion constructs are shown, with exon sequences in uppercase and intron sequences in lowercase and alternative sequences indicated. Red, U12-type consensus sequences; green, converted U2-type sequences; gray, NHE3 exon sequences; brown, linker sequences; orange, PEST sequences. The mutations converting cyan to yellow fluorescence are highlighted in cyan and yellow, respectively. The mutations introduced to change hydrophobic amino acids in the predicted transmembrane domain of exon 5 are in bold. (C) A plasmid containing the U2C and U12Y constructs in tandem (named U2C–U12Y) was created to ensure equal transfection of both constructs. The reciprocal plasmid (U12C–U2Y) with a reversed color scheme controlled for inherent differences in CFP and YFP fluorescence yield.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.