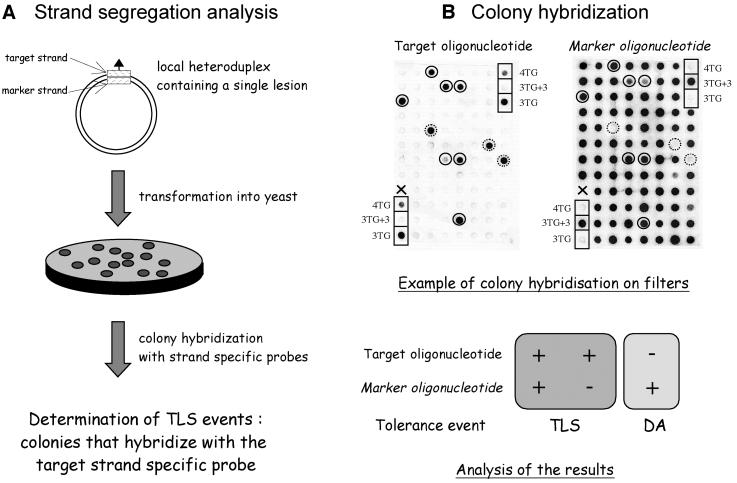
Fig. 1. Determination of TLS in vivo. Double-stranded plasmids carrying a single adduct (triangle), located within a short heteroduplex region (hatched rectangles), are constructed as described in Materials and methods, and used for strand segregation analysis as shown in (A). The sequence context of the heteroduplex region is shown in Figure 2 for the different lesion bypass assays implemented in the present paper. In (B), an example of a filter containing colonies obtained following transformation of plasmid pKB-3TG in wild-type yeast cells is shown. The left and right images are autoradiographs obtained upon hybridization with the target (3TG probe) and marker (3TG+3 probe) oligonucleotides, respectively. Control colonies transformed with plasmids pKB-4TG, pKB-3TG+3 and pKB-3TG are shown in boxes. Plasmid pKB-4TG is derived from plasmid pKB-3TG containing an additional T residue within sequence 5′-TTTG, thus mimicking the +1 mutation induced by AAF adducts in this sequence context (see Figure 2). As expected, the colonies containing control plasmid pKB-3TG+3 light up with probe 3TG+3 only. Conversely, colonies containing control plasmids pKB-3TG and pKB-4TG light up strongly and weakly with the 3TG probe only, respectively. Among the colonies analyzed, seven colonies (circles with solid outline) hybridize with both target and marker strand probes, suggesting that TLS occurred during replication. Three colonies (circles with broken outline) hybridize with target strand probe only, suggesting that TLS occurred during a gap-filling event. These latter TLS events most likely occur during gap filling of excision tracks generated during mismatch repair and have been shown to exhibit the same genetic requirements as TLS events that occur during replication (Baynton et al., 1998; data not shown). All other colonies responding to probe 3TG+3 only are scored as damage-avoidance events. A single colony marked with an X failed to grow (no signal with either probe).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.