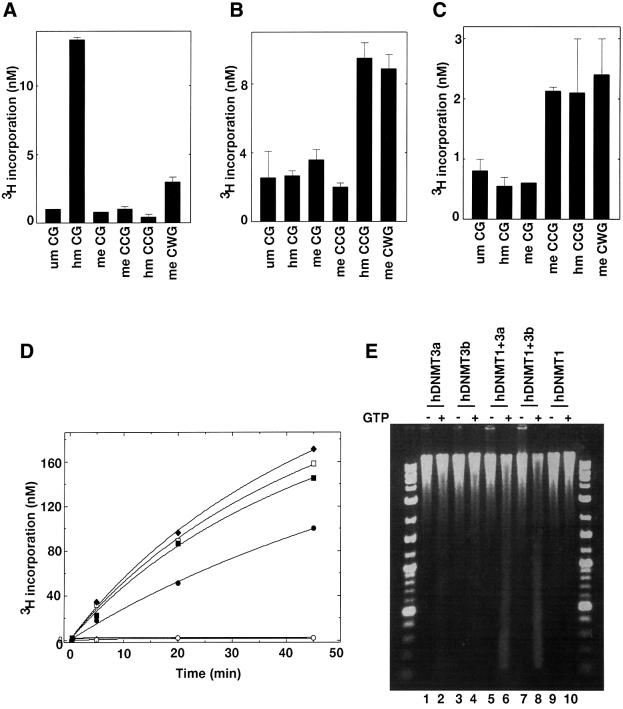
Fig. 6. Differential methylation of pre-methylated substrate DNA in vitro and methylation spreading in vivo. In vitro methylation using pre-methylated substrate with (A) hDNMT1, (B) hDNMT3a or (C) hDNMT3b. Equimolar concentrations of DNMTs were added to the reaction mix. Incorporation was measured after 30 min for hDNMT1 and 1 h for hDNMT3a or hDNMT3b. The experiment was conducted in duplicate and the average of two independent experiments is reported. (D) Co-operation for DNA methylation spreading in vitro. Reaction velocity curve of hDNMT1 and hDNMT3b (filled diamonds), hDNMT1, hDNMT3a and hDNMT3b (open squares), hDNMT1 and hDNMT3a (filled squares), hDNMT1 (filled circles), hDNMT3a (open circles), hDNMT3a and hDNMT3b (filled triangle), and hDNMT3b (open diamonds). Methylation by hDNMT3a, hDNMT3b and hDNMT3a, and hDNMT3b were similar and the data points are overlapping. The average values of two independent data points are plotted. The rates of methylation are shown in Table I. (E) Methylation spreading in vivo. Genomic DNAs from insect cells expressing different hDNMTs (indicated on top) were purified and digested with McrBC in the presence (+) or absence (–) of cofactor GTP. The digested DNA was analyzed on a 1.2% agarose gel. A 10 kbp to 100 bp ladder was used as a marker. um, unmethylated; hm, heminethylated; me, methylated.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.