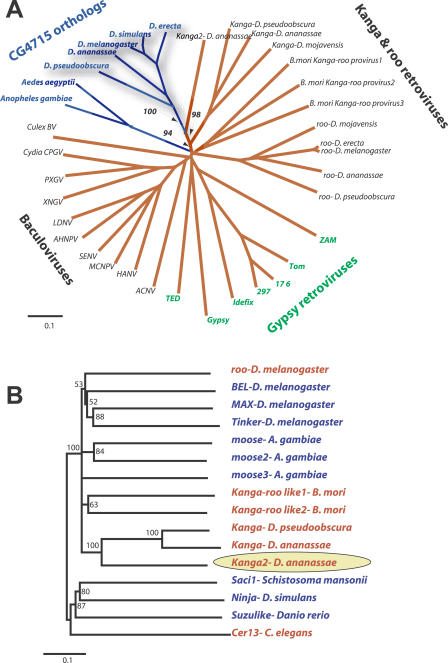
Figure 3. CG4715/Iris Relationships to Viral Envelope Genes.
(A) The central domains of CG4715 and related viral env genes were aligned, and a neighbor-joining phylogenetic tree constructed. The tree separates the CG4715-env superfamily into four groups: the baculoviruses, the BEL clade retroviruses roo and Kanga, the gypsy-like retroviruses, and host genome borne CG4715 orthologs in Drosophila and mosquito genomes. While the tree overall does not provide high resolution to discern the order of divergence of each of the clades, there is very strong phylogenetic resolution (bootstrap support of key nodes shown) to unambiguously group CG4715 orthologs with the Kanga retrovirus lineage, indicating that this lineage of retroviruses is the likely source of the CG4715 lineage.
(B) Neighbor-joining phylogeny of selected representatives from the BEL clade of retrotransposons indicates that the Kanga retroviruses from Drosophila genomes form a monophyletic clade (the presumed ancestor of CG4715 is indicated by a yellow oval). Most retrotransposons in the BEL lineage do not possess an env gene (blue lettering) while many elements that do (red) have acquired non-homologous env genes acquired from a different viral source [19,20].