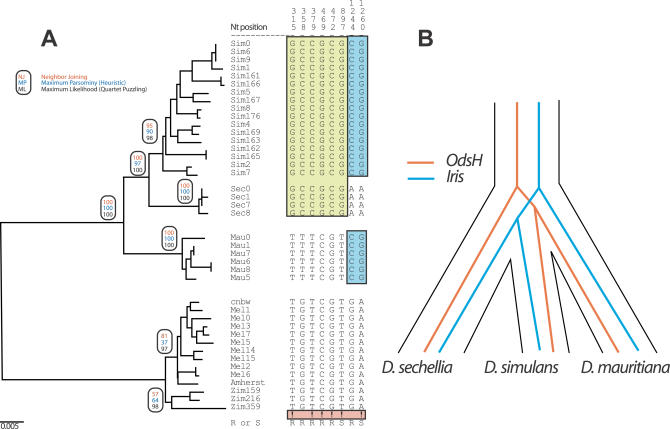
Figure 9. Iris Phylogeny in Closely Related Species.
(A) Phylogenetic analysis of Iris coding regions from different strains of D. melanogaster, D. simulans, D. sechellia, and D. mauritiana, the latter three species believed to have diverged less than half a million years ago [51]. Based on distance, parsimony or likelihood methods (bootstrap values indicated in ovals), the phylogeny clearly separates the three species. This is largely due to six sites that are “unambiguous” as far as phylogenetic information is concerned, indicated with “!.” An unambiguous site is defined as one in which the same derived nucleotide is found fixed in two of the three species (e.g., D. simulans and D. sechellia), whereas the third species (e.g., D. mauritiana) is fixed for the ancestral nucleotide, corresponding to the out-group, D. melanogaster.
(B) Iris is only the second known gene to inform about the phylogeny of the three sibling species D. simulans, D. sechellia, and D. mauritiana with statistical significance. In the Iris phylogeny, D. mauritiana branched earliest while previously, D. sechellia was found to branch earliest. This suggests that speciation events' chronology among these three species is more complicated than suggested previously [52].