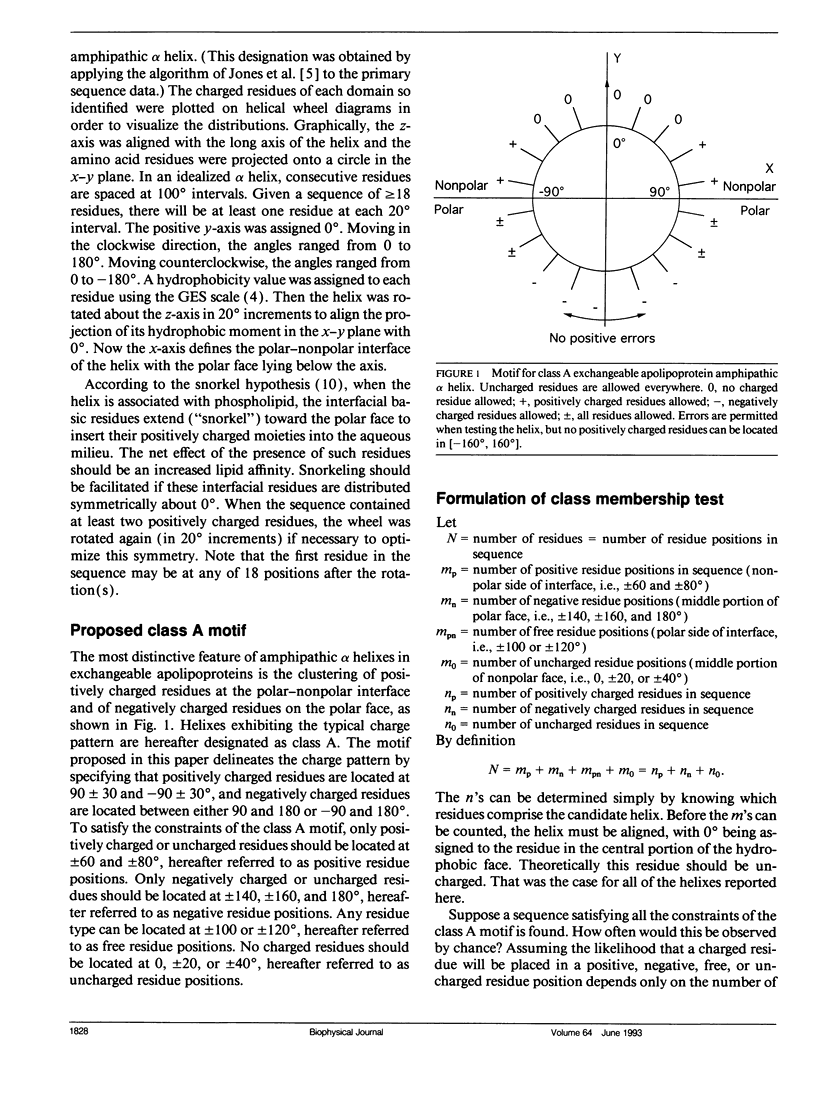
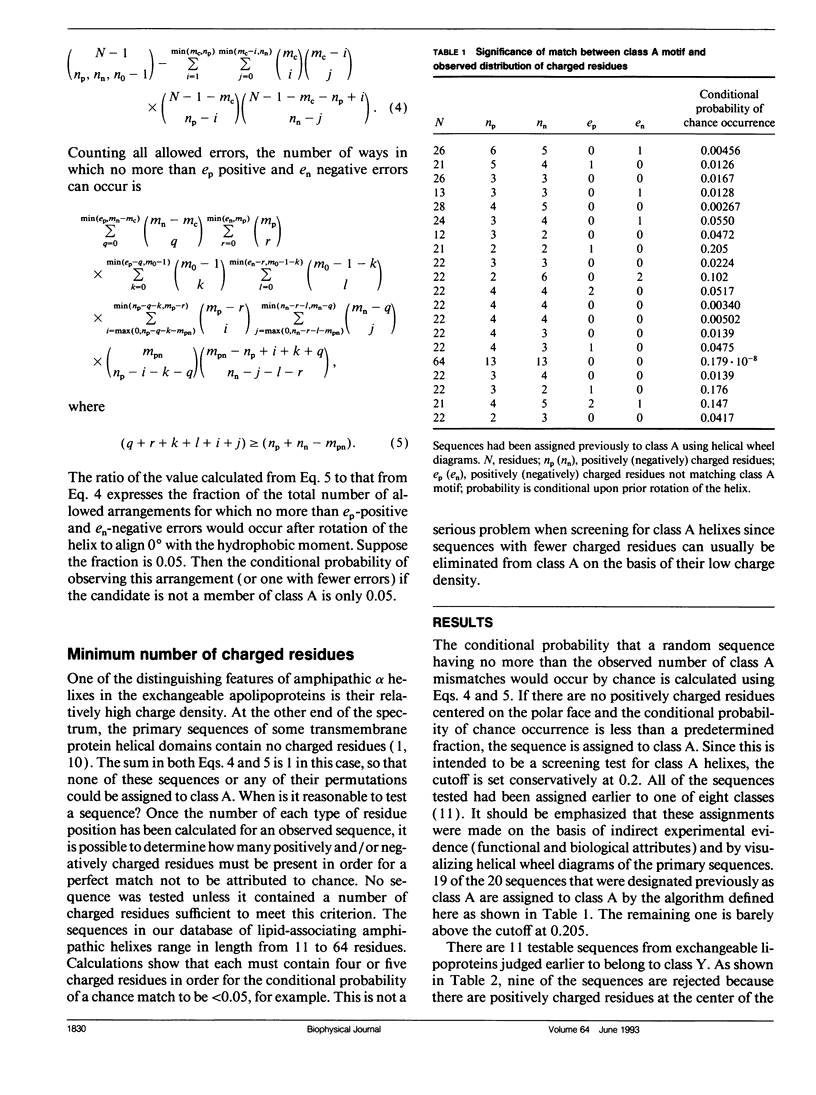
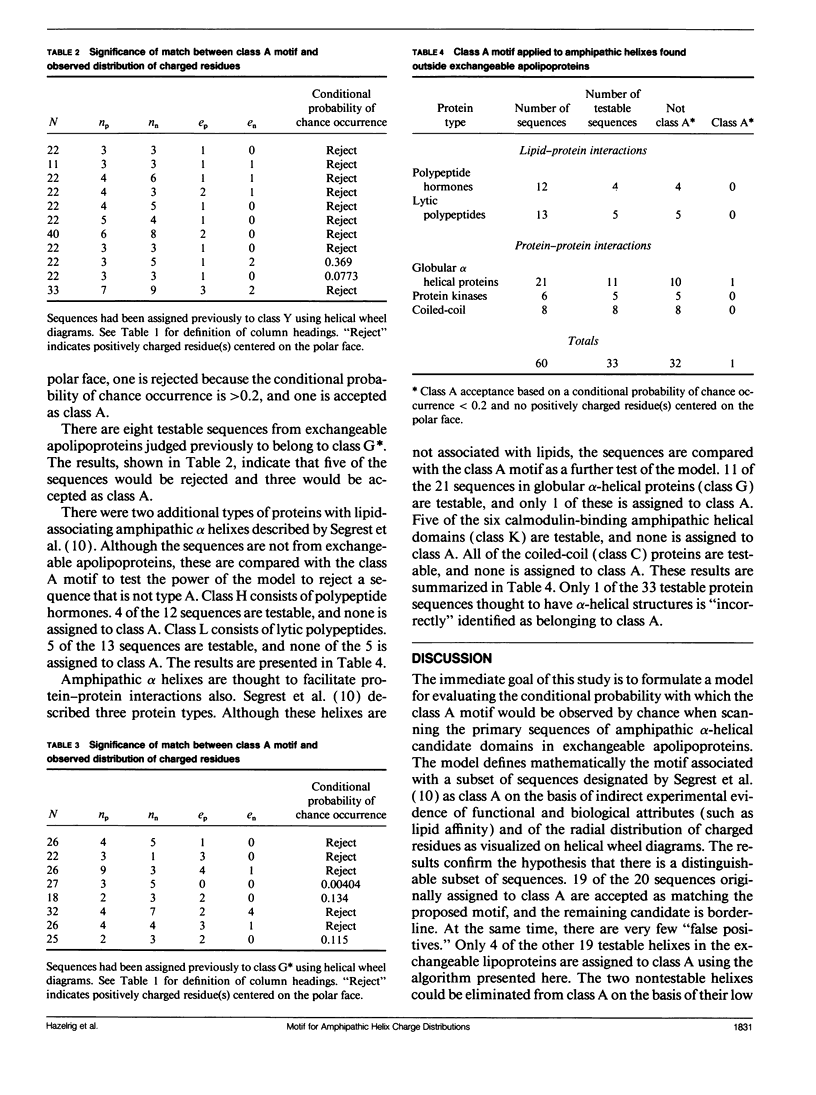
Abstract
Multiple amphipathic alpha-helical candidate domains have been identified in exchangeable apolipoproteins by sequence analysis and indirect experimental evidence. The distribution of charged residues can differ within and between these apolipoproteins. Segrest et al. (Segrest, J. P., H. DeLoof, J. G. Dohlman, C. G. Brouillette, and G. M. Anantharamaiah. 1990. Proteins. 8:103-117.) argued that these differences are correlated with lipid affinity. A mathematically defined motif for the particular charge distribution associated with high lipid affinity (class A) is proposed. Primary sequence data from protein segments proposed previously to have an amphipathic alpha-helical structure are scanned. Counting formulas are presented for determining the conditional probability that the match between an observed charge distribution and the proposed motif would occur by chance. Because the preselected helical segments are short (the modal length is 22) and the motif definition imposes multiple constraints on the acceptable distributions, the computer-based algorithm is quite feasible computationally. 19 of the 20 segments previously assigned to class A match the motif sufficiently well (the remaining one is borderline), while very few others "erroneously" pass the screening test. These results confirm the original assignments of the candidate domains and, thus, support the hypothesis that there is a distinguishable subset of helixes having high lipid affinity. This counting approach is applicable to a growing subset of protein sequence analysis problems in which the segment lengths are short and the motif is complex.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen J. P., Feher G., Yeates T. O., Komiya H., Rees D. C. Structure of the reaction center from Rhodobacter sphaeroides R-26: the protein subunits. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6162–6166. doi: 10.1073/pnas.84.17.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski M. S., Elshourbagy N., Taylor J. M., Gordon J. I. Comparative analysis of repeated sequences in rat apolipoproteins A-I, A-IV, and E. Proc Natl Acad Sci U S A. 1985 Feb;82(4):992–996. doi: 10.1073/pnas.82.4.992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman D. M., Steitz T. A., Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- Jones M. K., Anantharamaiah G. M., Segrest J. P. Computer programs to identify and classify amphipathic alpha helical domains. J Lipid Res. 1992 Feb;33(2):287–296. [PubMed] [Google Scholar]
- Kubota Y., Takahashi S., Nishikawa K., Ooi T. Homology in protein sequences expressed by correlation coefficients. J Theor Biol. 1981 Jul 21;91(2):347–361. doi: 10.1016/0022-5193(81)90237-x. [DOI] [PubMed] [Google Scholar]
- Lim V. I. Algorithms for prediction of alpha-helical and beta-structural regions in globular proteins. J Mol Biol. 1974 Oct 5;88(4):873–894. doi: 10.1016/0022-2836(74)90405-7. [DOI] [PubMed] [Google Scholar]
- Schiffer M., Edmundson A. B. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys J. 1967 Mar;7(2):121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segrest J. P., De Loof H., Dohlman J. G., Brouillette C. G., Anantharamaiah G. M. Amphipathic helix motif: classes and properties. Proteins. 1990;8(2):103–117. doi: 10.1002/prot.340080202. [DOI] [PubMed] [Google Scholar]
- Segrest J. P., Jackson R. L., Morrisett J. D., Gotto A. M., Jr A molecular theory of lipid-protein interactions in the plasma lipoproteins. FEBS Lett. 1974 Jan 15;38(3):247–258. doi: 10.1016/0014-5793(74)80064-5. [DOI] [PubMed] [Google Scholar]
- Segrest J. P., Jones M. K., De Loof H., Brouillette C. G., Venkatachalapathi Y. V., Anantharamaiah G. M. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J Lipid Res. 1992 Feb;33(2):141–166. [PubMed] [Google Scholar]


