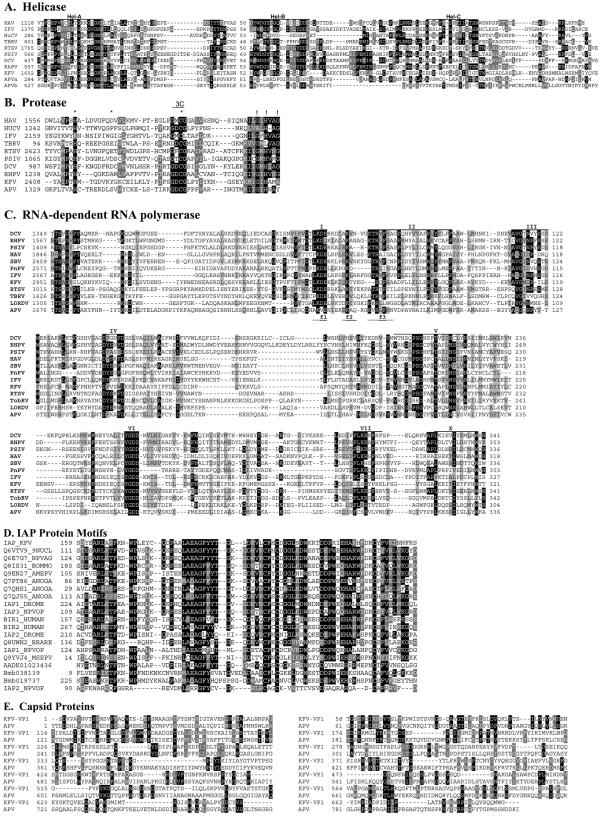
FIG. 3.
Comparison of the deduced amino acid sequences of the structural and nonstructural proteins of KFV and other picorna-like viruses. (A) Alignment of the conserved regions of the putative RNA helicase protein sequence from KFV with those of other picorna-like virus. The motifs recognized by Gorbalenya et al. (27) are labeled Hel-A, Hel-B, and Hel-C. (B) Alignment of KFV and picorna-like RNA virus chymotrypsin-related cysteine protease 3C-like protein sequences. The conserved residues of chymotrypsin-like proteases (26) that should form the catalytic triad are marked with asterisks, and putative substrate-binding residues are indicated by exclamation marks. The conserved catalytic cysteine motif associated with 3C proteases is indicated by the region labeled “3C.” (C) Alignment of KFV and picorna-like virus RdRp protein sequences. The amino acid sequences were aligned using the GAP program as described by Gorbalenya et al. (28). The motifs reviewed by Koonin and Dolja (36) are labeled I to VII. The f1 to f3 regions of the conserved motif F recognized by Bruenn (4) are denoted as such below the alignment sequences. (D) Comparison of the deduced amino acid sequences of representative BIR proteins of KFV, viral, dipteran, lepidopteran, and vertebrate origins. The KFV domain is labeled IAP_KFV. Other amino acid sequences are from UniProt (labeled with their entry numbers). The only exceptions are three possible novel IAPs from the Bombyx mori genome (64); these are predicted proteins Bmb038139 and Bmb019737, plus a partial sequence present in contig AADK01023436. Sequence identities of the sequences shown to the KFV sequence range from 58% identity (E = 7e−22) for the Choristoneura fumiferana defective nucleopolyhedrovirus (Q6VTV9_9NUCL) at the top down to ca. 30% for the IAP2 from the Orgyia pseudotsugata multicapsid polyhedrosis virus (IAP2_NPVOP). (E) Comparison of the deduced amino acid sequences of the structural proteins of KFV (VP1) and APV. Numbers to the initial left of alignments represent the residue numbers within the GenBank sequences. Numbers thereafter refer to residue distance from that point. Black shading indicates ≥50% identity; gray shading indicates ≥50% similarity. DCV, Drosophila C virus; RhPV, Rhopalosiphum padi virus; PSIV, Plautia stali intestine virus; PnPV, Perina nuda picorna-likevirus; HAV, human hepatitis A virus; RTSV, rice tungro spherical virus; HuCV, human calicivirus; TBRV and TobRV, tobacco ringspot virus.