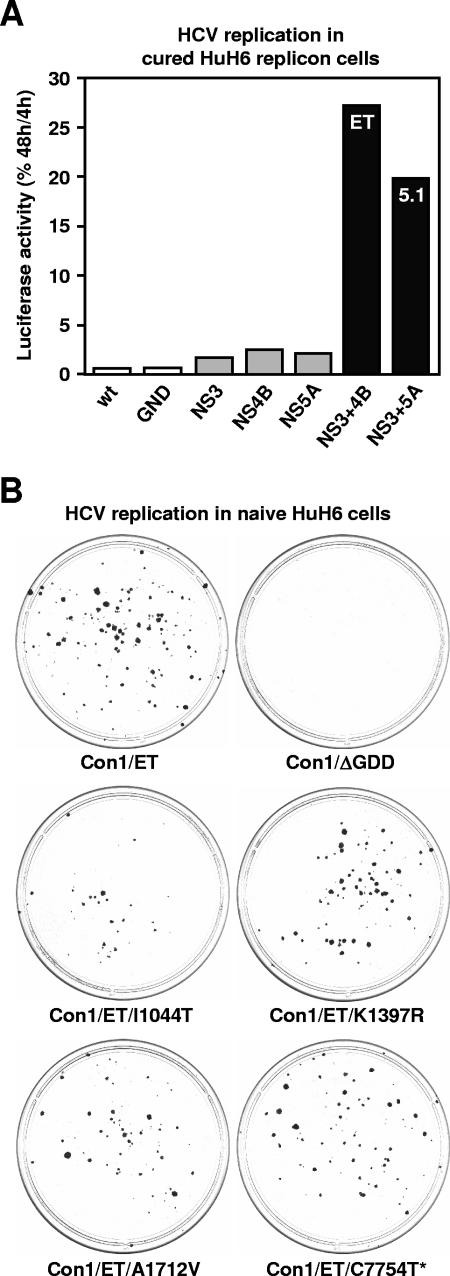
FIG. 2.
Effect of viral mutations on HCV RNA replication in HuH6 cells. (A) Effect of cell culture-adaptive mutations known to enhance Con1 replication in Huh-7 cells. Cured HuH6 replicon cells were transfected with in vitro-transcribed RNAs of I341PI-luc/NS3-3′ Con1 replicons and seeded into multiple cell culture dishes. After 4 and 48 h, cells were lysed and replication was quantified by measuring the luciferase activities. To correct for different transfection efficiencies, the 4-h values were set to 100%, and percentages of corresponding 48-h values were calculated. Column colors indicate the absence of cell culture-adaptive mutations (white), their presence in the coding sequence of a single (gray), or two different viral proteins (black). wt, wild-type Con1 sequence; GND, replicon with an inactivating mutation in the GDD motif of NS5B; NS3, replicon with E1202G and T1280I mutations in the protease domain of NS3; NS4B, replicon with K1846T mutation in the center of NS4B; NS5A, replicon with the S2204I mutation in NS5A; NS3+4B, replicon with the ET combination of mutations in NS3 and 4B (E1202G, T1280I, and K1846T); NS3+5A, replicon with the 5.1 combination of mutations in NS3 and 5A (E1202G, T1280I, and S2204I). The result of a single representative experiment is shown. (B) Colony formation assay to analyze the effect of conserved mutations that had been identified in HuH6 cells after the transfection of the Con1 replicon I389Neo/NS3-3′/ET. Naive HuH6 cells were electroporated with in vitro-transcribed replicons that contained either the ET combination of adaptive mutations, an inactivating mutation in the GDD motif of NS5B, or the ET combination of adaptive mutations plus one of the newly identified nucleotide substitutions: T3472C (I1044T in NS3), A4531G (K1397R in NS3), C5474G (A1712V in NS4A), or C7754U (silent mutation in NS5B). After the transfection, cells were seeded in 10-cm dishes, cultured for 4 weeks in the presence of 800-μg/ml G418, cultured for 2 additional weeks in the presence of 500-μg/ml G418, fixed, and stained with Coomassie blue. The positions of nucleotide/amino acid exchanges cited above refer to those in the Con1 consensus genome (EMBL database accession number AJ238799).