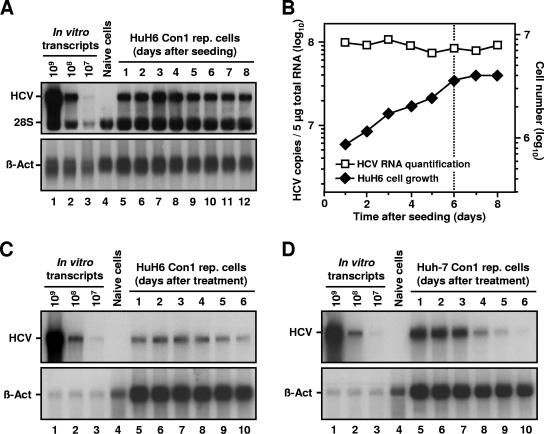
FIG. 4.
HCV RNA replication in nondividing HuH6 cells. (A) Detection of positive-stranded HCV RNAs in a growing culture of HuH6 Con1 replicon cells. Equal numbers of cells were seeded in multiple 10-cm cell culture dishes and harvested at given time points. Total RNA was prepared, and samples of 5 μg (each) were analyzed by Northern hybridization to a riboprobe complementary to the neo gene or to β-actin mRNAs (lanes 5 to 12). Different amounts of in vitro transcripts were used as standards (lanes 1 to 3). RNA isolated from naive HuH6 cells served as a negative control (lane 4). The positions of replicon RNA, 28S rRNA, and β-actin are indicated. (B) Comparison between the number of HCV RNAs (squares) and cell number at the time of harvest (diamonds). Prior to RNA preparation, HuH6 Con1 replicon cells were trypsinized and counted, and the number of cells per 10-cm cell culture dish was calculated. Hybridization signals of positive-stranded HCV RNAs and β-actin mRNAs shown in panel A were quantified and HCV RNA copy numbers were corrected for loading differences. Note that HCV copy and cell numbers are given in different log scales. The dotted line indicates the transition from the exponential to the stationary growth phase of the cell culture. (C and D) Detection of positive-stranded HCV RNAs in roscovitine-treated HuH6 Con1 cells and Huh-7 cells of clone 9-13, respectively. Equal numbers of cells were seeded in multiple 10-cm cell culture dishes, treated with 20 μM roscovitine, and harvested at given time points. Total RNA was prepared, and samples of 5 μg were analyzed by Northern hybridization as described above. The positions of replicon RNA and β-actin are indicated.