Fig. 1. Diagram of Kir3.
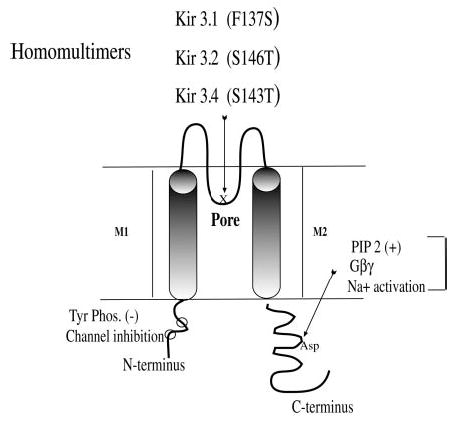
Kir3 channels share a common design characterized by a cytoplasmic N terminus (~90 amino acids) and C terminus with two transmembrane domains M1 and M2, which surround the ion-selective pore region (P or H5). Kir3 channels function as active heteromultimers (e.g. Kir3.1 pairs with other subtypes). Mutations in the P region enhance the activity of homomers. This diagram shows the P region with the site of specific point mutations that produce functional homomeric channels: Kir3.1(F137S), Kir3.2(S146T), and Kir3.4(S143T). The presumed PIP2-Na+ 1-Gβγ interaction site in the C terminus is indicated. Kir3 is also inhibited by tyrosine phosphorylation (11) with phosphorylation sites in the N terminus noted (○). Other sites of Gβγ interaction have been identified, but are not illustrated.
