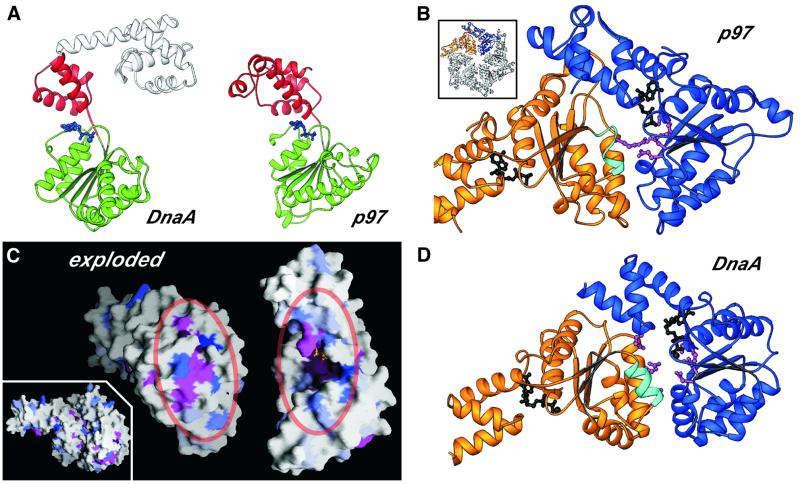
Fig. 4. (A) RIBBONS diagram of DnaA and p97 (residues 191–458) showing the high degree of structural conservation across the AAA+ domain of the two proteins (the r.m.s.d. between the two proteins is 3.2 Å over 192 Cα positions). (B) RIBBONS diagram of the AAA+ region of a p97 dimer excised from the hexameric structure (inset) (Zhang et al., 2000) depicting the typical oligomeric arrangement of AAA+ protomers. The ADP bound at the interface is shown in black. The helix containing the Box VII motif is shown in cyan and the key arginine residue present at the dimerization interface is depicted as a magenta ball-and-stick model. (C) Inset: surface depiction (Nicholls et al., 1991) of the DnaA dimer modeled on p97, showing the high degree of complementarity between the monomers. The structural alignment of the AAA+ regions was generated using least-squares fitting of the DnaA AAA+ domain on each of two neighboring p97 AAA+ domains from the p97 hexamer. The exploded view reveals the clustering of conserved residues at the dimerization interface to form the bipartite nucleotide-interaction site. The degree of conservation among all known DnaA sequences is indicated by the degree of blue shading (Figure 1B); invariant (magenta) and chemically conserved residues (pink) are also highlighted. (D) RIBBONS diagram of the DnaA AAA+ domain (residues D77S130–G290N348) model dimer shown in blue and gold. Critical elements present at the predicted dimer interface are highlighted as in Figure 4B.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.