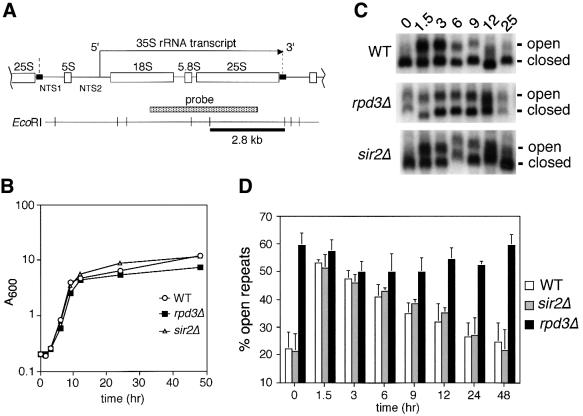
Fig. 1. Deletion of RPD3 prevents inactivation of rDNA repeats as cells exit log phase. (A) Schematic representation of the rDNA in S.cerevisiae, showing the initiation site and direction of 35S transcription by Pol I. The Pol III-transcribed 5S rRNA gene separates the non-transcribed spacer regions (NTS1 and NTS2). A Pol I enhancer element is located at the 3′ end of each 35S repeat (small black rectangles). The 2.8 kb EcoRI restriction fragment and probe are indicated. The boundaries of one rDNA repeat are indicated by vertical dashed lines. (B) Representative growth curve of yeast cultures used for the psoralen cross-linking assay. The strains used were JS311 (WT), JS490 (rpd3Δ) and JS218 (sir2Δ). (C) Gel retardation of actively transcribed rDNA genes through psoralen cross-linking. Aliquots harvested at the indicated time points were photoreacted with psoralen. Genomic DNA was digested with EcoRI and rDNA-specific fragments were detected using the 35S probe. For simplicity, only the 2.8 kb fragment is shown. The actively transcribed repeats (open) and inactive repeats (closed) are indicated on the right. (D) Quantitation of actively transcribed rDNA repeats. By PhosphorImager analysis, the percentage of the total rDNA repeats that were in the open chromatin conformation (slow migrating band) was calculated for each strain and time point. The average percentages from three independent experiments are shown in a bar graph along with standard deviation error bars.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.