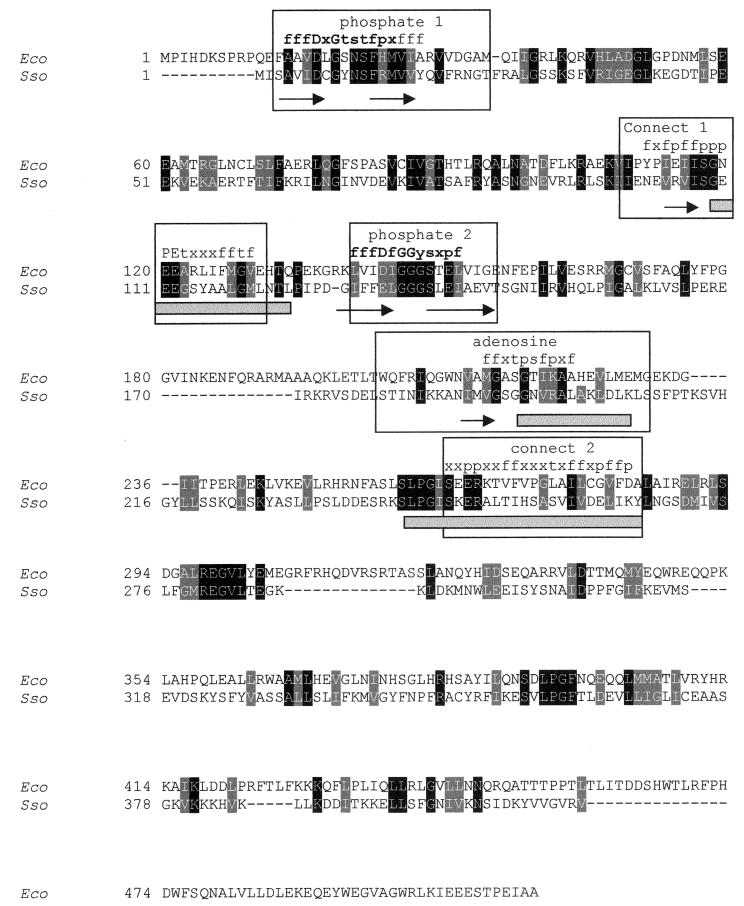
FIG. 2.
Pairwise alignment of the amino acid sequences of E. coli PPX and the putative PPX from S. solfataricus. The deduced amino acid sequence from S. solfataricus (EMBL accession no. CAC39441) (Sso) was aligned with the E. coli PPX sequence (NCBI protein database accession no. NP_416997) (Eco). Identical residues (black shading) and similar residues (gray shading) are indicated. The ATPase conserved motifs (6) are boxed, and within these boxes β strands are indicated by arrows and α helices are indicated by gray rectangles. The consensus sequence motif among 45 aligned PPXs is indicated over the alignment in terms of the following amino acids groups: f, partly hydrophobic (VLIFWYMCGATKHR); t, tiny (GSAT); s, small (GSATNDVCP); p, tiny plus polar (GSATNDQEKHR); and x, any amino acid. The residues in bold correspond to the consensus sequences described previously (6).