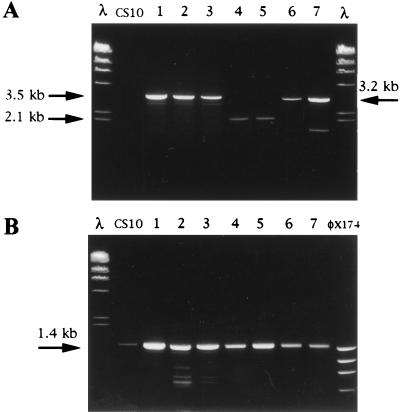
FIG. 6.
PCR analysis to detect site of integration in pAPGUSNP transformants. Template DNA for 5′ and 3′ nor-1 and pyrG integration assays was prepared with a rapid boiling procedure. (A) GUS+ and GUS− transformants (lanes 1 through 7) and CS10 (negative control) analyzed for 5′ nor-1, 3′ nor-1, or pyrG integration. Primers PCR3 were used to detect pyrG integration (lanes 1, 2, and 3; GUS− transformants) and generated a 3.5-kb PCR fragment. Primers PCR2 were used to detect 3′ nor-1 integration (lanes 4 and 5; GUS+ transformants) and generated a 2.1-kb PCR fragment. Primers PCR1 were used to detect 5′ nor-1 integration (lanes 6 and 7; GUS+ transformants) and generated a 3.1-kb PCR fragment. (B) DNA from all seven transformants and the control strain CS10 amplified with a primer pair specific for exon III of the versicolorin B synthase gene (positive control). A 1.4-kb PCR fragment was expected for all fungal isolates. Lanes containing molecular size markers in panels A and B are represented by λ HindIII digest and φx174 HaeIII digest. Nucleotide sequences of all primers are found in Table 1.