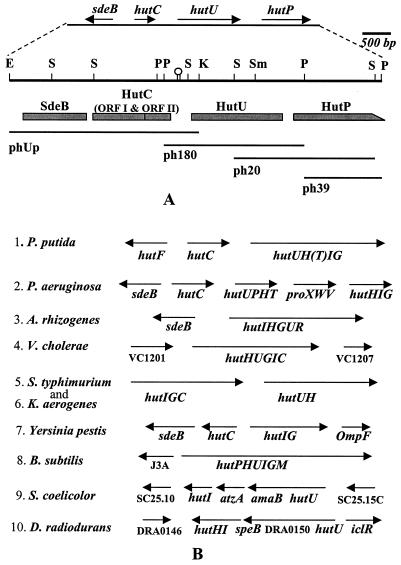
FIG. 1.
Physical map and the organization of the hutU region of P. syringae. (A) A 6.578-kbp DNA from the hutU region of P. syringae was sequenced and analyzed by cloning the region on four overlapping DNA restriction fragments (phUp, ph180, ph20, and ph39) shown below the physical map. Only the major restriction enzyme recognition sites (E, EcoRI; K, KpnI; P, PstI; S, SalI; Sm, SmaI) are indicated on the physical map. The genes (sdeB, hutC, hutU, and hutP) and their directions of transcription are shown at the top. The putative ORFs, such as SdeB, HutC ORFI and ORFII, HutU, and HutP, are shown as shaded boxes below the physical map. The incomplete ORF of HutP is indicated as a box with a slope at the C-terminal end. A hairpin structure between HutC and HutU indicates the location of a dyad structure in the DNA sequence. (B) The organization of the genes in the hutU region of a few bacteria identified either by earlier genetic studies or by recent genome sequence analyses is shown for comparison. The directions of transcription are indicated above the genes by arrows. The genes encoding various enzymes involved in histidine utilization (hut) are abbreviated as hutH (for histidase), hutU (for urocanase), hutI (for imidazolone propionate hydrolase), hutF (for FIGLUase), hutG (for formylglutamate amidohydrolase), hutT (for inducible histidine/urocanate transporter), hutP (putative transporter with similarity to purine-cytosine permease), hutC orfI (putative repressor of hut operon), and hutC orfII (3′ downstream ORF of hutC). The unknown ORFs of the hutU region in some bacteria are indicated directly by their ORF identification numbers (e.g., VC1201, VC1207, J3A, SC25.10, SC25.15C, and DRA0146).