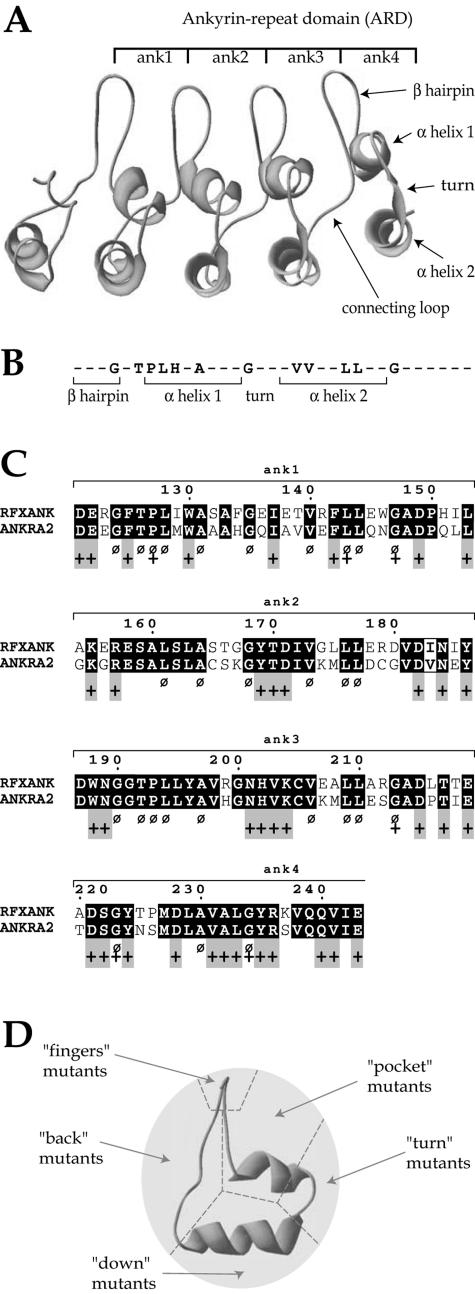
FIG. 3.
Selection of residues mutated in RFXANK. (A) Computer-generated three-dimensional model of the ankyrin repeat domain of RFXANK. The four ankyrin repeats are shown. Each consists of a β-hairpin and two anti-parallel α-helices. (B) Consensus ankyrin repeat sequence. Residues highly conserved between ankyrin repeat-containing proteins are indicated. Nonconserved residues are represented by dashes. (C) Alignment of the ankyrin repeat domains from RFXANK and ANKRA2. Residues conserved between the two proteins are highlighted in black. Residues that are highly conserved between many different ankyrin repeat proteins, and that were for this reason excluded from our selection, are indicated by a Ø sign. Residues predicted to be solvent accessible are indicated by a + sign. Gray boxes indicate residues that are accessible and conserved between RFXANK and ANKRA2 but not conserved in other ankyrin proteins. These residues were mutated in this study. (D) Residues selected for mutagenesis were divided into five groups according to their positions in the ankyrin repeat structure. A side view of the structure is shown. “Finger” mutants are located in the β-hairpin, “pocket” mutants in the ankyrin “groove,” “turn” mutants in the loop connecting the two α-helices, “down” mutants in the downward surface of the external α-helix, and “back” mutants in the loop connecting two adjacent ankyrin repeats.