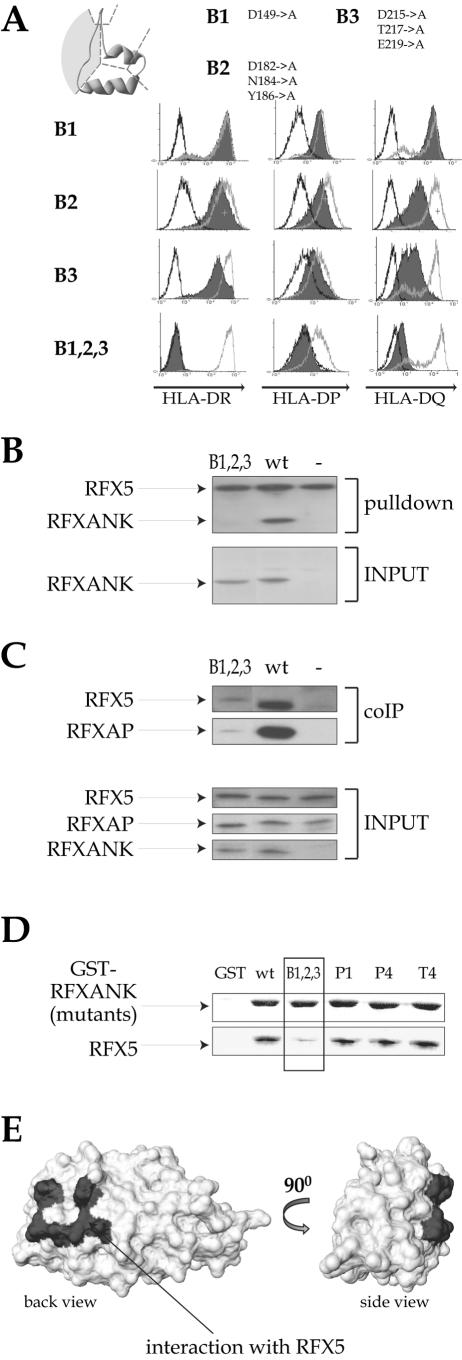
FIG.5.
Identification of the surface that mediates the interaction with RFX5. (A) Mutations located in the loops connecting two adjacent ankyrin repeats (“back” mutants) were analyzed in the complementation assay as described in the legend to Fig. 1B. (B) Promoter pull-down experiments with equal amounts of extracts prepared from BLS-1 cells complemented with either the B1,2,3 mutant or wild-type (wt) RFXANK were performed as described in the legend to Fig. 1C. Non complemented BLS-1 cells were used as a negative control (−). Input extracts were analyzed for the presence of RFXANK. The amounts of the other subunits of the RFX complex were equal in all input extracts (data not shown). (C) Vaccinia-T7-produced Flag-RFXANK proteins (wild-type or B1,2,3 mutant), RFX5 and RFXAP, were incubated together to form RFX complexes. Anti-Flag antibody was then used to immunoprecipitate RFXANK, and the presence of RFX5 and RFXAP in the immune complexes was analyzed by Western blotting. No RFXANK was added in the control lane (−). Inputs were analyzed in the bottom panels. (D) Equal amounts of glutathione-Sepharose bound GST or GST fusions containing wild-type and mutant RFXANK were incubated with in vitro-translated 35S-labeled RFX5 and washed. Bound proteins were fractionated by SDS-PAGE and analyzed by Coomassie blue staining (for GST fusions; top) or autoradiography (for RFX5; bottom). (E) Space-filling model of the RFXANK ARD. The RFXANK-RFX5 interaction domain is marked in gray.