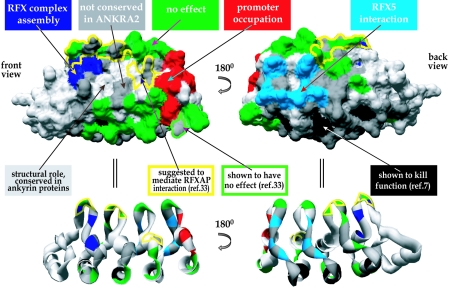
FIG. 8.
Model representing all known functions and interaction surfaces in RFXANK. Front and back views of the space-filling (top) and ribbon (bottom) models of RFXANK's ARD are marked with different colors representing different mutations and surfaces analyzed in this and previous studies. Residues implicated in RFX complex assembly (Fig. 6) are marked in dark blue, residues important for promoter occupation in vivo (Fig. 7) are marked in red, and those mediating interactions with RFX5 (Fig. 5) are marked in light blue. Residues at which the mutations had no effect (Fig. 4) are shown in green. Dark gray was used for residues which are not conserved in ANKRA2 and were therefore not analyzed. Light gray indicates residues which were not analyzed because they either are conserved in different ankyrin-containing proteins (and are therefore unlikely to mediate specific functions of RFXANK) or are inaccessible at the protein surface. Light gray was also used for the helix located adjacent to the ARD (leftmost part in the front view). Mutations of the residues marked in black were shown previously to be deleterious for RFXANK function (7). Since these mutations lie in the conserved α-helices, they probably disrupt the structure of the ARD. The yellow contour indicates residues suggested previously to mediate interactions with RFXAP (33). The same study showed that mutations of the residues marked with a green contour have no effect.