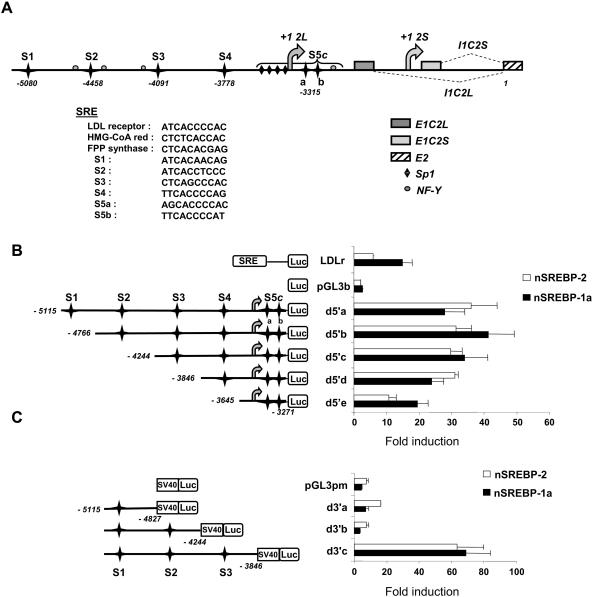
FIG. 1.
5′-flanking region of the human CASP-2 gene. (A) Sequence positions are numbered relative to the major translation initiation site (position 1) located in the first coding exon (E2) common to both caspase-2 isoforms. Both transcription initiation sites (+1 2L and +1 2S) and the corresponding first introns (I1C2L and I1C2S) are indicated. Gray boxes indicate the position of the first exons, specific for each Casp-2 mRNA. The six SRE sites are represented by black stars. Putative Sp1 and NF-Y sites are also shown. S5c designates a complex composed of four Sp1 sites, two juxtaposed SREs (S5a and S5b), and one NF-Y site. The sequence of each site is indicated, together with those of sites found in the promoters of bona fide SREBP target genes. (B and C) Each CASP-2 gene promoter fragment was prepared as described in Materials and Methods. Five-prime deletions (from d5′a to d5′e) (B) were subcloned into the pGL3b vector and 3′ deletions (from d3′a to d3′c) (C) in the pGL3 pm vector. Each SRE site is identified (S1 to S5c). HepG2 cells were transfected with each reporter vector (or with LDLr, empty pGL3b, or pGL3 pm vectors as controls) and cotransfected with expression vectors for the SREBP-1a (nSREBP-1a) or SREBP-2 (nSREBP-2) mature form. Luciferase activity was measured 24 h later and normalized to β-galactosidase activity. The bars represent inductions relative to basal luciferase activity without SREBP cotransfection. The values are the means (± standard deviations) of at least three independent experiments.