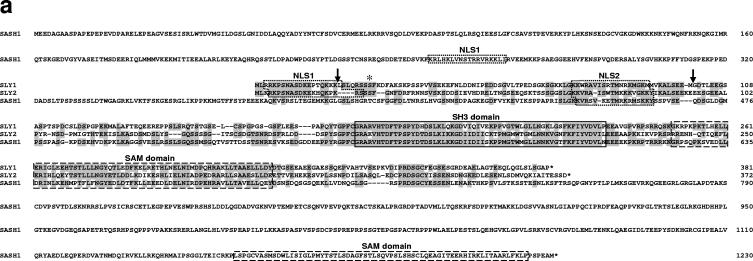
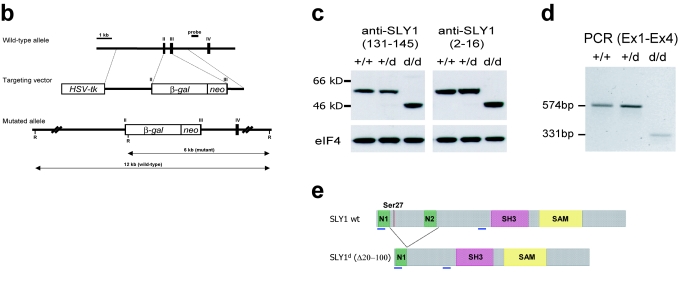
FIG. 1.
Generation of Sly1 mutant mice. (a) Amino acid sequence alignment of murine SLY1, SLY2, and SASH1 proteins is shown. Identical residues are shaded and protein domains are indicated by boxes. The arrows assign the deletion in SLY1d caused by skipping of exons 2 and 3. The asterisk marks phosphorylated Ser27. (b) The organization of part of the murine Sly1 genomic locus (top), the linearized targeting vector (middle), and the targeted Sly1 allele (bottom) are shown. The β-galactosidase cDNA and the neomycin resistance cassette were inserted between codon 26 (exon 2) and codon 72 (exon 3) of the Sly1 gene. The β-galactosidase cDNA contains a polyadenylation signal. Exon sequences are represented by solid rectangles, intron sequences by a black line (R, EcoRI). (c) Lysates of splenocytes from wild-type (+/+), heterozygous (+/d), and homozygous mutant (d/d) mice were analyzed by Western blot with two independent rabbit polyclonal antisera directed against amino acids 2 to 16 or 131 to 145 of the wild-type SLY1 protein. Antisera specific for eIF4 served as loading control. (d) The skipping of exons 2 and 3 was revealed by reverse transcription-PCR with exon 1- and exon 4-specific primers in splenic mRNA from wild-type (+/+) and heterozygous (+/d) as well as homozygous (d/d) mutant mice. (e) Schematic representation of the mutant SLY1d protein. N, nuclear localization signal; blue lines, immunogenic peptides for generation of SLY1-specific antisera.