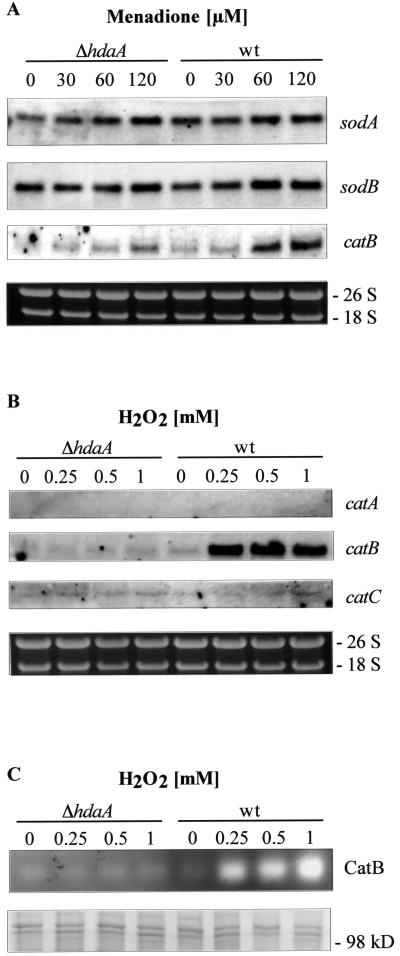
FIG. 5.
Regulation of detoxifying enzymes of the cellular antioxidant response in hdaA deletion strains. H4 as a ΔhdaA strain and H1 as a control representing the wt were used. (A) Transcription rates of the coding sequences of superoxide dismutases A and B and catalase B at various concentrations of menadione. Northern analyses were performed with 10 μg of total RNA isolated from strains grown for 18 h in minimal medium and for a further 3 h in the presence of the corresponding MD concentration. Transcripts were detected with digoxigenin-labeled probes specific for the respective sequences. (B) Transcription rates of three catalase genes (catA, catB, and catC) in the presence of different concentrations of H2O2. Strains were grown for 18 h in minimal medium and for a further 3 h in the presence of the corresponding H2O2 concentration. Ethidium bromide-stained rRNA was used as a quality and loading control in panels A and B. (C) CatB activity in protein extracts from H4 (ΔhdaA) and H1 (wt) growing under conditions as described for panel B. Protein extracts were loaded onto a native polyacrylamide gel, and CatB activities were determined by staining as described in Materials and Methods. As a loading control, aliquots used for the zymogram analysis were loaded onto an SDS-polyacrylamide gel and stained with Coomassie blue (lower panel).