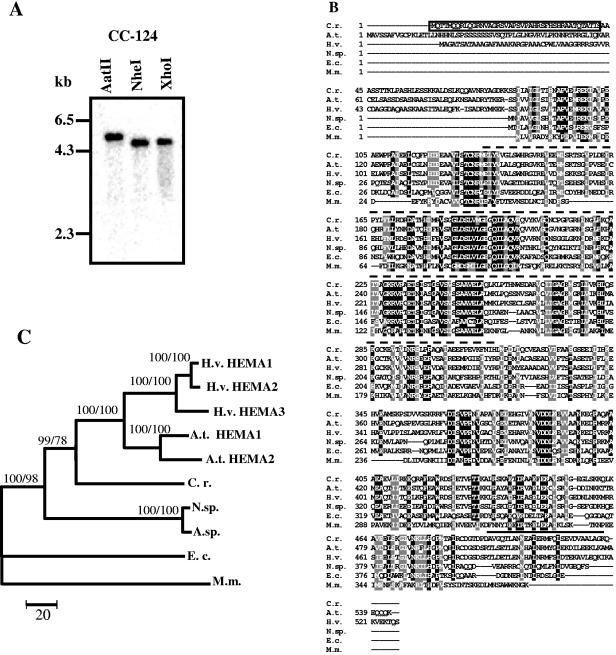
FIG. 1.
Test for the number of HEMA genes in the C. reinhardtii genome and relatedness of the C. reinhardtii HEMA product to those of other organisms. (A) Southern blot of C. reinhardtii genomic DNA. Genomic DNA (10 μg) from wild-type strain CC-124 was digested with either AatII, NheI, or XhoI. The sizes of marker fragments are shown on the left. (B) Multiple-sequence alignment of six HEMA proteins, generated by the ClustalW program. The deduced amino acid sequence of the C. reinhardtii HEMA (GenBank accession number AF305613) was compared with those of Arabidopsis thaliana (A.t. HEMA1; NP_176125), Hordeum vulgare (H.v. HEMA1; BAA25167), Nostoc sp. strain PCC7120 (N.sp; NP_485085), Escherichia coli K-12 (E.c; BAA36068), and Methanococcus maripaludis S2 (M.m; NP_987208). A black background indicates identical amino acids; residues highlighted with a gray background are similar. N/Q, D/E, R/K, S/T, F/Y, A/G, and V/I/L/M were considered similar. The transit peptide of C. reinhardtii HEMA, predicted by the ChloroP program (12), is boxed. The probe used for Northern blot and Southern blot analyses encodes the stretch of amino acids indicated by a dotted line above the sequences. A coiled-coil motif predicted for the C-terminal end is marked by a solid line above the sequences (6, 31). (C) The phylogenetic tree is based on the alignment of proteins described above and the following additional proteins: Arabidopsis thaliana HEMA2 (NP_172465), Hordeum vulgare HEMA2 (CAA60055) and HEMA3 (BAA25168), and Anabaena sp. (A.sp.; AAB58164). The phylogenetic tree was obtained by MEGA 2.1 distance-neighbor joining based on the number of amino acid substitutions. Bootstrap support is from consensus trees after 500 repetitions. HEMA sequences from E. coli and Methanococcus maripaludis were taken as outgroups. Distances are indicated by the scale bar. Numbers at nodes indicate bootstrap support values (first number from distance-neighbor joining, second from maximum parsimony).