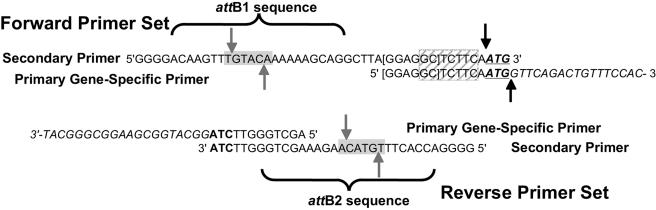
FIG. 1.
Diagram of the nested PCRs. The gene-specific forward primer (shown here for gene SMa1985) included the first 20 bases of the ORF (in italics) with an ATG start codon (in boldface, underlined). The ribosome-binding sequence (brackets) added to the ORF and sequences between the ribosome-binding sequence and the start codon generate a SapI restriction site (hatched gray box) that generates a consistent overhang around the start codon (black solid arrows). The gene-specific reverse primer included the last 20 bases of the ORF (in italics) and an ATC sequence (in boldface) that will generate TAG at the end of the ORF to insert an amber suppressible stop codon. The secondary forward and reverse primers included the attB1 and attB2 sequences, respectively, as indicated by the brackets below the sequence and a Bsp1407I restriction site (gray-shaded box, gray solid arrows).