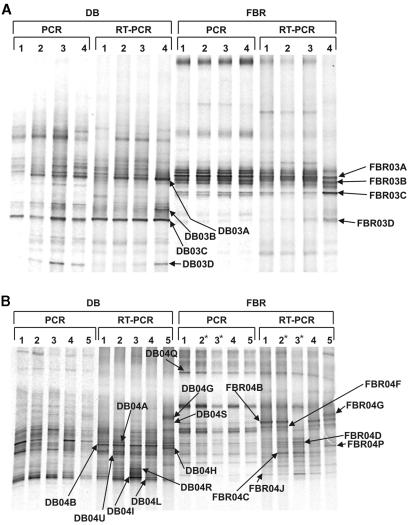
FIG. 1.
(A) DT-DGGE analysis of DB and FBR samples following anoxic, nitrate-amended incubations (December 2003). Samples were incubated for 10 h in degassed nitrate-amended marine medium. PCR (DNA) and RT-PCR (RNA) treatments were supplemented with the following sulfide concentrations: lanes 1, original (nonincubated) biofilter sample; lanes 2, 0 μM H2S; lanes 3, 500 μM H2S; lanes 4, 5,000 μM H2S. (B) DT-DGGE analysis of DB and FBR samples following anoxic, nitrate-amended, and oxic incubations (March 2004). Samples were incubated for 20 h in degassed nitrate-amended marine medium (anoxic) or aerated medium (oxic). PCR (DNA) and RT-PCR (RNA) treatments were supplemented with the following sulfide concentrations: lanes 1, original (nonincubated) biofilter sample; lanes 2, 1,000 μM H2S; lanes 3, 5,000 μM H2S; lanes 2*, 100 μM H2S; lanes 3*, 1,000 μM H2S; lanes 4, 1,000 μM S2O3 (anoxic); lanes 5, 1,000 μM S2O3 (oxic). Asterisks indicate sulfide concentration discrepancies for FBR treatments.