Abstract
Gene expression changes of glutamate-producing Corynebacterium glutamicum were identified in transcriptome comparisons by DNA microarray analysis. During glutamate production induced by a temperature shift, C. glutamicum strain 2262 showed significantly higher mRNA levels of the NCgl2816 and NCgl2817 genes than its non-glutamate-producing derivative 2262NP. Reverse transcription-PCR analysis showed that the two genes together constitute an operon. NCgl2816 putatively codes for a lactate permease, while NCgl2817 was demonstrated to encode quinone-dependent l-lactate dehydrogenase, which was named LldD. C. glutamicum LldD displayed Michaelis-Menten kinetics for the substrate l-lactate with a Km of about 0.51 mM. The specific activity of LldD was about 10-fold higher during growth on l-lactate or on an l-lactate-glucose mixture than during growth on glucose, d-lactate, or pyruvate, while the specific activity of quinone-dependent d-lactate dehydrogenase differed little with the carbon source. RNA levels of NCgl2816 and lldD were about 18-fold higher during growth on l-lactate than on pyruvate. Disruption of the NCgl2816-lldD operon resulted in loss of the ability to utilize l-lactate as the sole carbon source. Expression of lldD restored l-lactate utilization, indicating that the function of the permease gene NCgl2816 is dispensable, while LldD is essential, for growth of C. glutamicum on l-lactate.
Corynebacterium glutamicum is a gram-positive bacterium that was originally isolated as an l-glutamate producer from soil samples in the early 1950s (18, 40). It belongs to a broad, diverse group of mycolic acid-containing bacteria that share the property of having an unusual cell envelope composition and architecture, differing from those of other gram-positive bacteria (5, 31). Since its discovery, C. glutamicum has been used for the production of monosodium glutamate, a flavoring agent, with a current annual production scale of approximately 1,500,000 tons (11).
Glutamate production by C. glutamicum is characterized by a low pentose phosphate pathway flux (22); low conversion of 2-oxoglutarate to oxaloacetate in the tricarboxylic acid cycle (22), likely due to low activity of the oxoglutarate dehydrogenase complex (3, 12, 37, 38); pyruvate carboxylase fulfilling the anaplerotic demand, since phosphoenolpyruvate carboxylase is inhibited by glutamate (2, 28); and glutamate excretion by a specific export system (5, 9, 12).
A peculiarity of glutamate production is that it has to be elicited by external triggers, e.g., by addition of penicillin or detergents, by biotin limitation, or by ethambutol addition (5,17, 20, 33, 37, 38). In some strains glutamate production can be induced by a temperature shift (3). This is the case with C. glutamicum 2262. However, the production properties of this strain are unstable, and upon prolonged continuous cultivation, a non-glutamate-producing mutant, referred to below as C. glutamicum 2262NP, was isolated (41). The temperature shift was found to induce a metabolic transition in C. glutamicum 2262 that was not observed in the mutant C. glutamicum 2262NP. This transition typically involves a decrease in the activities of the 2-oxoglutarate dehydrogenase complex and of pyruvate dehydrogenase, a redirection of the 2-oxoglutarate flux toward glutamate production, and the formation of l-lactate as a by-product (41).
Since glutamate production by C. glutamicum has not yet been analyzed with respect to global gene expression changes, we employed DNA microarray analysis for transcriptome comparisons between glutamate-producing C. glutamicum 2262 and its non-glutamate-producing mutant 2262NP. Moreover, the existence of this pair of strains, which grow very similarly in continuous cultivation and differ in their glutamate-producing capacity, provided an opportunity to determine gene expression changes specific for glutamate production under the defined conditions of a chemostat. Here we characterize an operon, induced during temperature-triggered glutamate production, that is necessary for the (re)utilization of l-lactate.
(Part of the work described here belongs to the planned dissertation of C. Stansen at the Faculty of Mathematics and Natural Sciences of the Heinrich-Heine-Universität, Düsseldorf, Germany.)
MATERIALS AND METHODS
Bacterial strains, medium composition, and cultivation conditions.
Bacterial strains and plasmids used are listed in Table 1. Wild-type C. glutamicum ATCC 13032 as well as (for temperature-triggered glutamate production) C. glutamicum 2262 and its non-glutamate-producing mutant 2262NP were used (41). The chemostat medium was prepared as described previously (41). Polypropylene glycol (1.3 g liter−1) was used as an antifoaming agent. For shake flask cultivations, the minimal medium CgXII (16) supplemented with 0.03 g liter−1 protocatechuate and the complex medium BHI (Difco) were used. For chemostat cultures, the inoculum was grown in shake flask culture at 33°C in modified MCGC medium (41), supplemented with NaHPO4 (3.8 g liter−1) and urea (0.39 mg liter−1). The glucose concentration was increased to as much as 34 g liter−1, and the pH was set at 7.6 with NaOH. A 50-ml volume of this overnight culture was used to inoculate a 3-liter bioreactor (APPLIKON, Holland) containing 1.5liters of modified MCGC. When glucose was exhausted, the culture was supplemented continuously with modified MCGC. At the onset of supplementation, the culture temperature was increased from 33 to 39°C. The pH set point was 7.6, regulated with 12 N NH3. The agitation was set at 1,200 rpm to avoid oxygen limitation.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| C. glutamicum | ||
| ATCC 13032 | Biotin-auxotrophic wild type | ATCC |
| 13032::pK18mobNCgl2816int | NCgl2816 inactivation mutant of ATCC 13032 | This work |
| 2262 | Glutamate-producing strain | 41 |
| 2262NP | Non-glutamate-producing mutant of 2262 | 41 |
| E. coli DH5α | supE44 ΔlacU169 (φ80dlacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Invitrogen |
| Plasmids | ||
| pK18mob | Kanr; vector for integration into C. glutamicum; (pK18 oriVEc lacZα) | 35 |
| pK18mobNCgl2816int | pK18mob carrying an internal fragment of NCgl2816 | This work |
| pEKEx3 | Specr; C. glutamicum/E. coli shuttle vector for regulated gene expression (Ptac lacq pBL1 oriVCg pUC18 oriVEc) | E. Radmacher and L. Eggeling |
| pEKEx3-lldD | pEKEx3 carrying the NCgl2816/lldD gene | This work |
Determination of metabolite concentrations.
During cultivation, samples were collected to determine biomass, glucose, glutamate, and l-lactate concentrations. The cell concentration was determined by absorbance at 570 nm. After centrifugation of the sample (13,000 × g, 5 min), the concentrations of l-lactate and glucose were determined enzymatically (Roche, Germany). Intracellular glutamate concentrations and glutamate concentrations in the culture supernatants were determined as described previously (41).
Quinone-dependent l-lactate dehydrogenase assay.
For determination of enzyme activities, exponentially growing cells were harvested by centrifugation (4,500 × g, 5 min, 4°C) and washed twice with 50 mM ice-cold KH2PO4, pH 7.0. Cell extracts were prepared directly or after storage at −70°C by resuspension in 1 ml of 50 mM KH2PO4, pH 7.0, and disruption by ultrasonic treatment at 4°C (UP 200S; Dr. Hielscher GmbH, Teltow, Germany) at an amplitude of 55% and a duty cycle of 0.5 for 6 min. After centrifugation at 4°C for 60 min at 13,000 × g, enzyme activity was determined immediately in the cell-free supernatant. l-Lactate dehydrogenase activity was determined by a modified assay according to reference 23. Reaction mixtures of 1 ml contained 100 mM KH2PO4 (pH 7.5)-50 μM 2,6-dichloroindophenol (DCPIP) and 20 μl crude extract. Quinone-dependent l-lactate dehydrogenase was assayed spectrophotometrically at 30°C by determining the decrease in absorbance of DCPIP (ɛ600 = 20 cm2 μmol−1). The reaction was started by addition of 20 mM l-lactate, 10 mM d-lactate, or 20 mM racemic malic acid.
Comparative genome and transcriptome analysis using DNA microarrays.
Generation of C. glutamicum DNA microarrays, total RNA preparation, cDNA synthesis, preparation and labeling of genomic DNA, DNA microarray hybridization, and statistical data analysis were performed as described previously (14, 21, 29, 42).
Construction of plasmids and strains.
The genome sequence of C. glutamicum deposited in GenBank as accession number NC_003450 was used. Plasmids were constructed in Escherichia coli DH5α from PCR-generated fragments (Expand High Fidelity PCR kit; Roche Diagnostics) by using C. glutamicum ATCC 13032 genomic DNA prepared according to the method of Eikmanns et al. (6) as a template. E. coli was transformed by the RbCl2 method (10), while C. glutamicum was transformed via electroporation (39). All transformants were analyzed by plasmid analysis and/or PCR with appropriate primers, respectively. To construct an NCgl2816 inactivation mutant of C. glutamicum ATCC 13032, an internal 468-bp fragment was PCR amplified using primers NCgl2816-int_fw (5′-GCAACGAGTTCCTCTCCACAATC-3′ [the nucleotide corresponding to nucleotide {nt} 3118442 of NC_003450 is underlined]) and NCgl2816-int_rev (5′-ACAGGCCGATGAACACTAGGAG-3′ [the nucleotide corresponding to nt 3118909 of NC_003450 is underlined]). Subsequently, this fragment was blunted and cloned into the SmaI site of pK18mob (35). Gene inactivation with pK18mobNCgl2816int was carried out as described previously (27a). The correct genotype of the inactivation mutant was verified by PCR analysis using primer NCgl2816-int_proof_fw (5′-TGACCATGAGGTTGGGCCAATC-3′ [the nucleotide corresponding to nt 3118215 of NC_003450 is underlined]) and the universal primer M13-24F (on pK18mob). In order to construct pEKEx3-lldD, lldD was amplifed by PCR using primers lldD_fw (5′-AAGGAGATATAGATATGGTGAAACGTCAACTGCC-3′ [the nucleotide corresponding to nt 3119621 of NC_003450 is underlined, and italicized nucleotides correspond to the ribosome binding site of T7 gene 10]) and lldD_rev (5′-TTAAATCTCCGCCGCTGC AGAACGAGTT-3′ [the nucleotide corresponding to nt 3120883 of NC_003450 is underlined]). The PCR fragment was blunted and cloned into the SmaI site of pEKEx3.
Reverse transcription-PCR (RT-PCR) analysis.
RNA was prepared as described previously (14, 21, 29, 42) and incubated in 0.1 M sodium acetate, pH 3.0, 5 mM MgSO4, and 0.3 U μl−1 DNase I (Roche Diagnostics, Mannheim, Germany) for 30 min at 37°C. After heat inactivation for 5 min at 75°C and phenol-chloroform extraction, the RNA was precipitated with LiCl as described elsewhere (34). After denaturation for 5 min at 65°C, reverse transcription of 350 ng RNA was performed with Omniscript Reverse Transcriptase (QIAGEN, Hilden, Germany) according to the manufacturer's instructions by using primer lldD_rev (described in the preceding section) for reaction cDNA-A and primer NCgl2816-int_proof_rev (5′-TGCTGCTCCGACTTGGTAACC-3′ [the nucleotide corresponding to nt 3119433 of NC_003450 is underlined]) for reaction cDNA-B. Subsequently, the cDNAs were amplified using combinations of the above-mentioned primers NCgl2816-int_proof_fw, NCgl2816-int_proof_rev, lldD_fw, and lldD_rev. As a control, dnaE RNA was reverse transcribed using RT-dnaE-rev (5′-CTGGAACCATGTCGTCCTAGAG-3′ [the nucleotide corresponding to nt 2251091 of NC_003450 is underlined]) and amplified using primers RT-dnaE-fw (5′-TGCCCTTCCGGCGATGTCCAA-3′ [the nucleotide corresponding to nt 2251460 of NC_003450 is underlined]) and RT-dnaE-rev.
RESULTS
Continuous cultivation of glutamate-producing C. glutamicum 2262 and its non-glutamate-producing mutant 2262NP.
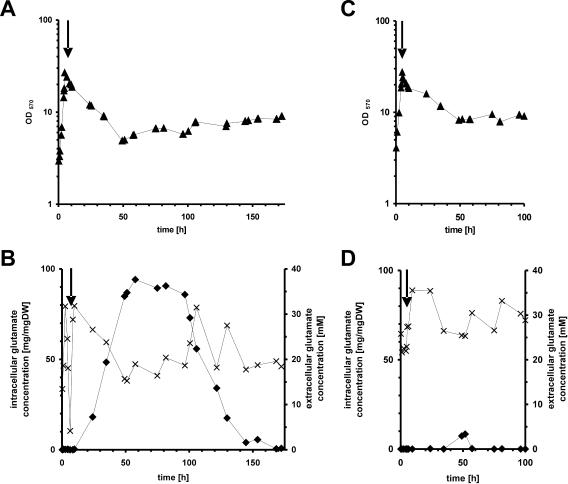
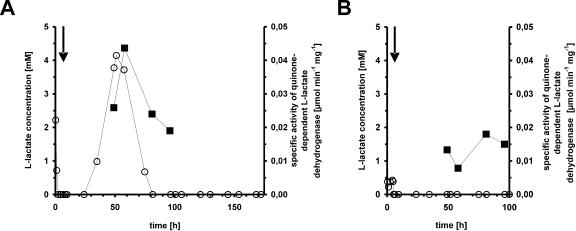
C. glutamicum 2262 produces l-glutamate in a temperature-triggered process. This process is known to be unstable, and a non-glutamate-producing mutant strain, C. glutamicum 2262NP, was isolated (41). In order to characterize gene expression patterns distinguishing these strains, continuous cultures were performed and samples were withdrawn for RNA isolation and DNA microarray analysis at 70 h. Figure 1A shows the continuous culture of C. glutamicum 2262. Due to the instability of this strain, the temperature shift from 33 to 39°C to induce glutamate production was imposed at the same time as the change from the initial batch mode to continuous cultivation with a dilution rate of 0.05 h−1. After this shift, C. glutamicum 2262 showed rapid growth reduction, while from 50 h of cultivation onward, the growth rate remained unchanged. As observed previously (41), glutamate accumulated in the culture medium shortly after the temperature increase, and maximal glutamate concentrations (>30 mM) were observed in the interval from 50 to 100 h of cultivation (Fig. 1B). l-Lactate accumulated transiently around 60 h of cultivation up to a concentration of 4.5 mM (Fig. 2A). The non-glutamate-producing strain 2262NP was cultivated under the same conditions (Fig. 1C). Concentrations of glutamate in the medium were below 5 mM, and those of l-lactate were below 0.5 mM (Fig. 1D and 2B).
FIG. 1.
Continuous culture of glutamate-producing C. glutamicum 2262 (A and B) and its non-glutamate-producing mutant 2262NP (C and D) at 39°C and a dilution rate of 0.05 h−1. (A and C) Optical densities at 570 nm (▴); (B and D) intracellular (×) and extracellular (⧫) glutamate concentrations. Arrows indicate the time when both the continuous supplementation of medium and the temperature increase from 33 to 39°C were imposed. The pH was maintained at 7.
FIG. 2.
l-Lactate concentrations in supernatants (○) and specific activities of quinone-dependent l-lactate dehydrogenase (▪) in crude extracts of glutamate-producing C. glutamicum 2262 (A) and of its non-glutamate-producing mutant 2262NP (B) during continuous culture. Samples analyzed were taken from the continuous cultures described in the legend to Fig. 1.
Transcriptome comparison of glutamate-producing C. glutamicum 2262 and its non-glutamate-producing mutant 2262NP.
DNA microarray technology has been established for C. glutamicum, with whole-genome microarrays based on PCR products obtained from genomic DNA of C. glutamicum ATCC 13032 (29, 42). To test whether these DNA microarrays are suitable for experiments with C. glutamicum 2262, genome comparisons between this strain and C. glutamicum ATCC 13032 were first performed as described previously for isogenic variants of C. glutamicum ATCC 13032 (42). Genomic DNAs of both strains were isolated and fluorescently labeled red and green, respectively, before simultaneous hybridization to the C. glutamicum ATCC 13032 DNA microarrays. While the genome comparisons revealed 333 genes in 26 loci that are present in C. glutamicum ATCC 13032 but are apparently lacking in C. glutamicum 2262, hybridization signals were observed for about 87% of the genes with genomic DNAs from both strains (see Fig. S1 in the supplemental material). A similar analysis revealed that 9 genes of 2,262 are deleted in the genome of 2262NP (see Table S1 in the supplemental material). Thus, genome and transcriptome comparisons of C. glutamicum 2262 can be performed using the C. glutamicum ATCC 13032 DNA microarrays.
The transcriptomes of glutamate-producing C. glutamicum 2262 and the non-glutamate-producing strain C. glutamicum 2262NP were compared using the samples withdrawn during the chemostat cultivations at 70 h of cultivation (see Fig. 1). Three independent DNA microarray experiments based on different RNA isolations were performed. These revealed that only two genes showed an mRNA level significantly (P < 0.05) changed by at least fourfold. NCgl2816 and NCgl2817 showed 11- and 9-fold-higher mRNA levels, respectively, in C. glutamicum 2262 than in C. glutamicum 2262NP. The two genes are adjacent to each other on the C. glutamicum chromosome, and their organization suggests that they are cotranscribed. The NCgl2816 gene encodes a putative transport protein and is situated upstream of NCgl2817, which putatively encodes a quinone-dependent l-lactate dehydrogenase. This led to the hypothesis that NCgl2817 and NCgl2816 constitute an operon for uptake and oxidation of l-lactate.
To follow up this hypothesis, samples were withdrawn from the chemostat cultures to assay for quinone-dependent l-lactate dehydrogenase in crude extracts. Whereas the glutamate-producing strain showed a transient increase in the specific activity of quinone-dependent l-lactate dehydrogenase, peaking at 45 nmol mg−1 min−1 (Fig. 2), this was not the case with the non-glutamate-producing strain 2262NP, where the activity did not exceed 20 nmol mg−1 min−1 (Fig. 2). Thus, the increased expression of NCgl2817 was reflected in an increased quinone-dependent l-lactate dehydrogenase activity. Importantly, the increase in the l-lactate concentration preceded the increase in the specific activity of quinone-dependent l-lactate dehydrogenase (Fig. 2). The l-lactate concentration in the culture medium decreased more rapidly than that calculated on the basis of the dilution kinetics of the continuous cultivation, indicating reutilization of l-lactate by C. glutamicum. These observations and the fact that, for thermodynamic reasons, quinone-dependent l-lactate dehydrogenase (EC 1.1.2.3; encoded by NCgl2817, lldD) operates in the l-lactate oxidation direction (1) are commensurate with the view that this enzyme is involved in reutilization of excreted l-lactate.
Inactivation of NCgl2816 in C. glutamicum ATCC 13032.
To study the function of the putative lactate utilization genes, the genetically tractable wild-type strain C. glutamicum ATCC 13032 was employed (15). The inactivation mutant 13032::pK18mobNCgl2816int was constructed and analyzed in order to determine whether the NCgl2816 gene is involved in l-lactate utilization. This mutant grew like the parent strain with pyruvate as a carbon source but was not able to utilize l-lactate (data not shown). Both the mutant and the parent strain grew similarly, albeit poorly, on d-lactate (data not shown).
In order to test whether the altered growth of the mutant is due to inactivation of the NCgl2816 gene or to polar effects on the downstream gene NCgl2817, the latter gene was cloned into pEKEx3 and expressed in the inactivation mutant to yield C. glutamicum 13032::pK18mobNCgl2816int(pEKEx3-NCgl2817). This recombinant was able to utilize l-lactate again (data not shown), demonstrating that only the function of NCgl2817 is essential for growth of C. glutamicum on l-lactate as the sole carbon source.
Properties and carbon source-dependent regulation of quinone-dependent l-lactate dehydrogenase from C. glutamicum ATCC 13032.
To test whether NCgl2817 encodes quinone- dependent l-lactate dehydrogenase, the specific activity of this enzyme was determined under various conditions. Only specific activities of quinone-dependent l-lactate dehydrogenase lower than 0.03 μmol mg−1 · min−1 were observed for strains 13032::pK18mobNCgl2816int and ATCC 13032 during growth on d-lactate or glucose (Table 2), and both strains showed specific activities of quinone-dependent l-lactate dehydrogenase of 0.02 μmol mg−1 · min−1 during growth on pyruvate. However, during growth on a mixture of l-lactate and glucose, C. glutamicum ATCC 13032 exhibited a high specific activity of 0.23 μmol mg−1· min−1, while that of 13032::pK18mobNCgl2816int was lower than 0.03 μmol mg−1 · min−1 (Table 2 and data not shown). Overexpression of NCgl2817 in ATCC 13032 and 13032::pK18mobNCgl2816int increased the specific activity of quinone-dependent l-lactate dehydrogenase during growth on glucose 47- and 29-fold, respectively (Table 2). Taking these data together, NCgl2817 is the gene encoding quinone-dependent l-lactate dehydrogenase, and we therefore renamed NCgl2817 as lldD.
TABLE 2.
Specific activities of quinone-dependent l-lactate dehydrogenase and quinone-dependent d-lactate dehydrogenase in various C. glutamicum strains during growth on glucose or l-lactate
| C. glutamicum strain | Sp act (μmol mg−1 · min−1) of the following enzyme during growth on the indicated substratea
|
|||
|---|---|---|---|---|
|
l-Lactate dehydrogenase (quinone dependent)
|
d-Lactate dehydrogenase (quinone dependent)
|
|||
| Glucose | l-Lactate | Glucose | l-Lactate | |
| ATCC 13032 | 0.03 | 0.23 | 0.13 | 0.19 |
| ATCC 13032(pEKEx3) | 0.03 | 0.20 | 0.11 | 0.25 |
| ATCC 13032(pEKEx3-lldD) | 1.41 | 0.99 | 0.09 | 0.15 |
| 13032::pK18mobNCgl2816int | 0.02 | NGb | 0.14 | NGc |
| 13032::pK18mobNCgl2816int(pEKEx3) | 0.02 | NG | 0.17 | NG |
| 13032::pK18mobNCgl2816int(pEKEx3-lldD) | 0.58 | 0.63 | 0.09 | 0.15 |
NG, no growth occurred on l-lactate.
C. glutamicum 13032::pK18mobNCgl2816int grown on a mixture of 100 mM l-lactate with 50 mM glucose showed a specific activity of quinone-dependent l-lactate dehydrogenase of 0.03 μmol mg−1 · min−1, while C. glutamicum ATCC 13032 showed 0.23 μmol mg−1 · min−1.
During growth on the l-lactate-glucose mixture, the specific activities of quinone-dependent d-lactate dehydrogenase were 0.15 μmol mg−1 · min−1 in
C. glutamicum 13032::pK18mobNCgl2816int and 0.20 μmol mg−1 · min−1 in C. glutamicum ATCC 13032.
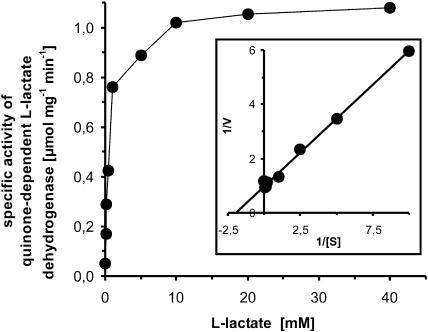
To estimate the substrate spectrum and the substrate affinity of C. glutamicum quinone-dependent l-lactate dehydrogenase (LldD), crude extracts of C. glutamicum ATCC 13032 overexpressing lldD and of a control strain carrying the empty vector were assayed for quinone reduction with various concentrations of l-lactate, d-lactate, and malate. The specific activity of l-lactate-dependent quinone reduction increased due to lldD overexpression, while malate-dependent or d-lactate-dependent quinone reduction did not. Michaelis-Menten kinetics were observed for l-lactate, and it was determined that 0.51 mM of l-lactate resulted in half-maximal enzyme activity (Fig. 3). The observed Vmax was 1.02 μmol mg−1 · min−1.
FIG. 3.
Substrate dependence of C. glutamicum WT(pEKEx3-lldD) quinone-dependent l-lactate dehydrogenase. Shown are specific activities of quinone-dependent l-lactate dehydrogenase in crude extracts from C. glutamicum WT(pEKEx3-lldD) with varying concentrations of the substrate l-lactate. (Inset) Double-reciprocal plot of the data.
To determine whether quinone-dependent l-lactate dehydrogenase is subject to carbon source regulation, the specific activity of this enzyme was assayed in crude extracts of C. glutamicum ATCC 13032 grown on different carbon sources. During growth on pyruvate, d-lactate, or glucose, the specific activity of quinone-dependent l-lactate dehydrogenase was lower than 0.03 μmol mg−1 · min−1. However, when l-lactate was the sole carbon source or when C. glutamicum ATCC 13032 grew on a mixture of glucose and l-lactate, high specific activities (0.23 μmol mg−1 · min−1) were observed. Thus, the amount of active enzyme is subject to carbon source regulation. The observed increase in the mRNA level of Ngl2817, as well as the increase in the specific activity of quinone-dependent l-lactate dehydrogenase, in glutamate-producing C. glutamicum 2262 suggests that the observed carbon source regulation occurs either by control of RNA stability or, more likely, at the transcriptional level.
Comparative transcriptome analysis of l-lactate- and pyruvate-grown C. glutamicum ATCC 13032.
We compared the transcriptomes of pyruvate- and l-lactate-grown cultures to test whether l-lactate-grown C. glutamicum ATCC 13032 cells showed high lldD mRNA levels and to determine whether NCgl2816 and lldD were the only genes specifically induced by l-lactate. The DNA microarray experiment revealed higher mRNA levels of NCgl2816 and lldD during growth on l-lactate than during growth on pyruvate (Table 3). Although NCgl2816 and lldD showed the highest increases in their mRNA levels on l-lactate compared to pyruvate, l-lactate utilization was characterized by additional gene expression changes. With l-lactate as the carbon source, expression of 20 further genes was more than fourfold higher, and expression of 10 genes more than fourfold lower, than for pyruvate-grown cultures (Table 3). The genes showing higher expression during growth on l-lactate mostly code for hypothetical proteins—putative regulatory, transport, and secretory proteins. Pyruvate-grown cells, on the other hand, showed higher expression of the pta-ack operon, coding for phosphotransacetylase (sixfold [Table 3]) and acetate kinase (twofold), and about fivefold-higher expression of aceA, coding for the glyoxylate cycle enzyme isocitrate lyase, and of the sdhCAB operon, coding for the tricarboxylic acid cycle enzyme succinate dehydrogenase (Table 3). These genes were proposed to belong to the regulon of the regulator of acetate metabolism RamB (7, 8). During growth on l-lactate, expression of genes of the iron starvation stimulon (19), like that of genes encoding proteins for the uptake and utilization of iron siderophores or heme, increased as follows: NCgl0377, 5.2-fold; NCgl0378, 1.8-fold; NCgl0380, 2.2-fold; NCgl0381, 2.1-fold; NCgl0382, 1.6-fold; NCgl0482, 4.2-fold; NCgl0483, 2.3-fold; NCgl0636, 2.1-fold; NCgl0637, 2.5-fold; NCgl0639, 3.6-fold; NCgl0773, 3.0-fold; NCgl00774, 3.3-fold; NCgl0776, 2.7-fold; NCgl0777, 1.9-fold; NCgl0778, 2.1-fold; NCgl00779, 2.9-fold; NCgl2970, 2.6-fold (see also Table 3). The genes for an AraC- and an ArsR-like regulator (NCgl0430 and NCgl0943) showed 4.2- and 10.1-fold-higher RNA levels, respectively, than during growth on pyruvate (Table 3).
TABLE 3.
Comparative gene expression analysis of C. glutamicum ATCC 13032 grown on pyruvate or l-lactate as the sole carbon source
| Genea | Annotationa | Relative mRNA levelb |
|---|---|---|
| NCgl0009 | Putative transcriptional regulator | 5.9 |
| NCgl0014 | Hypothetical membrane protein | 4.1 |
| NCgl0062 | Putative chloride channel protein | 4.0 |
| NCgl0122 | Hypothetical protein | 10.2 |
| NCgl0123 | Hypothetical protein | 7.6 |
| NCgl0359 | Succinate dehydrogenase, membrane anchor (sdhC) | 0.2 |
| NCgl0360 | Succinate dehydrogenase, flavoprotein (sdhA) | 0.2 |
| NCgl0377 | Putative heme transport-associated membrane protein | 5.2 |
| NCgl0430 | ArsR-type regulator | 4.2 |
| NCgl0482 | Putative iron siderophore ABC uptake system, ATPase | 4.2 |
| NCgl0943 | Transcriptional regulator, AraC family | 10.1 |
| NCgl1170 | Putative methylmalonyl coenzyme A epimerase | 0.2 |
| NCgl1262 | 3-Isopropylmalate dehydratase, large subunit (leuC) | 0.2 |
| NCgl1263 | 3-Isopropylmalate dehydratase, small subunit (leuD) | 0.2 |
| NCgl1470 | Putative protein kinase | 0.2 |
| NCgl1472 | Putative methylmalonyl coenzyme A mutase, beta subunit | 0.2 |
| NCgl1502 | Predicted iron-regulated ABC-type transporter SufB | 4.0 |
| NCgl1646 | Putative secreted hydrolase | 7.0 |
| NCgl1647 | Putative secreted protein | 5.0 |
| NCgl2248 | Isocitrate lysase (aceA) | 0.2 |
| NCgl2319 | Catechol 1,2-dioxygenase | 0.2 |
| NCgl2353 | Dipeptide-binding protein | 4.1 |
| NCgl2450 | Hypothetical protein | 6.7 |
| NCgl2451 | Hypothetical protein | 4.5 |
| NCgl2657 | Phosphotransacetylase (pta) | 0.2 |
| NCgl2713 | Putative transport protein | 5.4 |
| NCgl2719 | Putative ferredoxin/ferredoxin-NADP reductase | 4.2 |
| NCgl2816 | Putative transport protein | 17.9 |
| NCgl2817 | Quinone-dependent l-lactate dehydrogenase (lldD) | 17.4 |
| NCgl2965 | Putative transport protein | 4.6 |
Gene numbers and annotations of the revised C. glutamicum genome published by NCBI as NC_003450.
Ratio of the mRNA level in l-lactate-grown cells to that in pyruvate-grown cells. Statistically significant changes of at least fourfold in gene expression determined in three independent experiments from independent cultivations (P < 0.05 by Student's t test) are listed.
Operon structure of the NCgl2816-lldD genes.
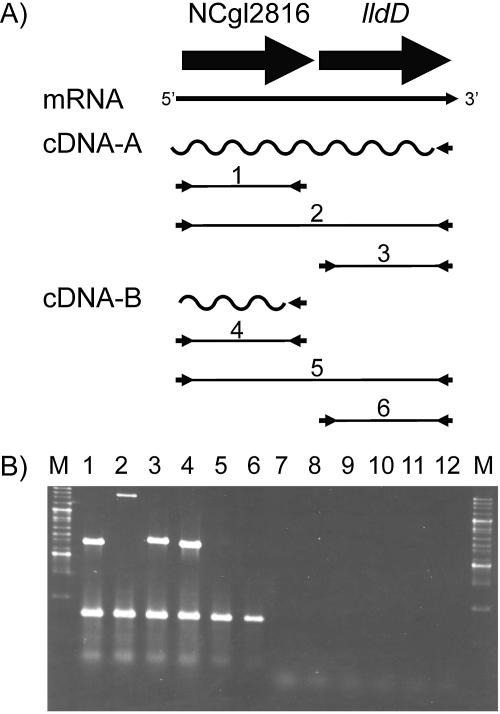
The transcriptional organization of the locus comprising the NCgl2816 and lldD genes was analyzed by RT-PCR (Fig. 4) (see also Materials and Methods). Total RNA isolated from C. glutamicum ATCC 13032 was transcribed into cDNA by using two different primers (RT-A and RT-B) in reverse transcriptase reactions A and B. The resulting products were then used for PCR amplifications 1 to 6 (Fig. 4A). As shown in Fig. 4B, a cDNA created with a primer annealing at the 3′ end of lldD (RT reaction cDNA-A) allowed not only the amplification of an lldD fragment (reaction 3) but also that of an NCgl2816 fragment (reaction 1). Further evidence was obtained that NCgl2816 and lldD are cotranscribed, since PCR amplification using primers annealing at the 5′ end of NCgl2816 and at the 3′ end of lldD yielded a PCR product covering both genes. As an internal control in the RT-PCR assays, we used dnaE, which encodes a subunit of DNA polymerase. Besides control reactions 5 and 6, where neither an lldD nor an NCgl2816-lldD product could be obtained with a cDNA covering only NCgl2816 (cDNA-B), six additional control reactions (reactions 7 to 12) were performed; these were identical to reactions 1 to 6, respectively, except that reverse transcriptase was omitted from the initial reactions. The fact that no PCR products were obtained in these reactions confirmed that the RNA was not contaminated with chromosomal DNA.
FIG. 4.
Transcriptional organization of the NCgl2816-lldD locus in C. glutamicum analyzed by RT-PCR. (A) Scheme showing the NCgl2816-lldD locus in C. glutamicum and the RT-PCRs used to determine cotranscription of NCgl2816 and lldD. RNA from wild-type C. glutamicum was transcribed into cDNA with two different primers in the two separate reverse transcriptase reactions cDNA-A and cDNA-B. Subsequently, these cDNAs were used as templates for the PCRs labeled 1 to 6. (B) Results from the RT-PCR analyses described above. The lower DNA fragment visible in lanes 1 to 6 represents dnaE, and RT-PCR of dnaE served as a positive control in all reactions. The upper bands correspond to the products of PCRs 1 to 6, diagramed in panel A. Lanes 7 to 12 represent control reactions confirming the absence of DNA in the RNA preparation. The reactions were identical to PCRs 1 to 6 (for which results are shown in lanes 1 to 6, respectively) except that reverse transcriptase was omitted in reactions cDNA-A and cDNA-B.
DISCUSSION
In this study, C. glutamicum lldD (NCgl2817) was demonstrated to encode the only l-lactate dehydrogenase essential for growth of C. glutamicum on l-lactate, as evidenced by the fact that an inactivation mutant of the NCgl2816-lldD operon was not able to utilize l-lactate unless lldD was expressed from a plasmid. The enzyme encoded by lldD is a quinone-dependent l-lactate dehydrogenase (EC 1.1.2.3). It is specific for l-lactate reduction and accepts neither d-lactate nor malate as a substrate. The Km of 0.51 mM for l-lactate is between that of the yeast enzyme (0.03 mM [44]) and that of the enzyme from the fungus Rhizopus oryzae (3.85 mM [30]). The specific activity of 0.23 μmol (mg protein)−1 min−1 determined in crude extracts of C. glutamicum ATCC 13032 grown on l-lactate agrees well with the calculated l-lactate uptake rate of about 0.11 μmol (mg [dry weight])−1 min−1 during consumption of this substrate (data not shown). Since biochemical data on bacterial quinone-dependent l-lactate dehydrogenases and the crystal structure of the yeast enzyme (44) have identified flavin mononucleotide as a prosthetic group, it is likely that the C. glutamicum enzyme also contains bound flavin mononucleotide as a cofactor.
Quinone-dependent l-lactate dehydrogenase from C. glutamicum belongs to the cluster of orthologous groups COG1304 and shows 88% sequence identity to Corynebacterium efficiens CE2762. The related mycobacteria possess two genes for quinone-dependent l-lactate dehydrogenases (lldD1 and lldD2). Sequence similarities of C. glutamicum LldD to the LldD2 enzymes from Mycobacterium tuberculosis (57%) (Rv1872c), Mycobacterium avium subsp. paratuberculosis (58%) (MAP1585c), Mycobacterium bovis (58%) (Mb1903c), and Mycobacterium leprae (58%) (ML2046) are higher than those to LldD1 enzymes from these mycobacteria (31% each for M. tuberculosis Rv0694, M. avium subsp. paratuberculosis MAP4154, and M. bovis Mb0713) and higher than those to LldD from E. coli (34%) and l-lactate ferricytochrome c reductase (29%) (Cyb2p) from the yeast Saccharomyces cerevisiae.
Besides quinone-dependent l-lactate dehydrogenase (EC 1.1.2.3), the following enzymes are known to occur in Corynebacterianeae: l-lactate monooxygenase (EC 1.13.12.4), NAD-dependent l-lactate dehydrogenase (EC 1.1.1.27), and quinone-dependent d-lactate dehydrogenase (EC 1.1.1.28). C. glutamicum possesses quinone-dependent d-lactate dehydrogenase (EC 1.1.1.28) activity likely to be encoded by NCgl0865 (dld) (1). An l-lactate monooxygenase (EC 1.13.12.4) has been characterized in Mycobacterium smegmatis as a flavoprotein utilizing molecular oxygen to oxidize l-lactate to acetate, CO2, and water (24). However, there are no indications that the genomes of C. glutamicum, C. efficiens, M. tuberculosis, M. leprae, and E. coli contain genes for l-lactate monooxygenases. On the other hand, genes for NAD-dependent l-lactate dehydrogenases (EC 1.1.1.27) are present in Corynebacterineae. These enzymes catalyze the exergonic NAD-dependent reduction of pyruvate to l-lactate and thus operate in the reverse direction from quinone-dependent lactate dehydrogenases, which catalyze the highly exergonic menaquinone-dependent oxidation of lactate (1). In C. glutamicum, ldhA (NCgl2810) encodes a NAD-dependent l-lactate dehydrogenase and is crucial for l-lactate production (13).
Recently, a membrane proteomics study identified the quinone-dependent l-lactate dehydrogenase LldD as a membrane-associated protein in C. glutamicum (36). Membrane association of LldD either could facilitate oxidation of l-lactate immediately after its uptake or, as proposed recently (25), could function in NADH oxidation. In a type II NADH dehydrogenase gene (ndh) disruption mutant of C. glutamicum KY9714, membrane fractions showed increased l-lactate oxidase activity, while ndh overexpression led to decreased l-lactate as well as malate oxidase activities (25). The authors suggested that coupling between cytoplasmic NAD-dependent l-lactate dehydrogenase and membrane-associated quinone-dependent l-lactate dehydrogenase and/or coupling between cytoplasmic NAD-dependent malate dehydrogenase and membrane-associated malate:quinone oxidoreductase results in effective NADH oxidation when type II NADH dehydrogenase function is defective (25). It remains to be studied whether double or triple mutants lacking ndh and either the l-lactate or the malate oxidation cycle or both cycles exhibit impaired growth relative to that of the parent strain.
NCgl2816 is transcribed together with lldD in an operon and codes for a putative permease of the major facilitator superfamily (TC 2.A.1) (43). Disruption of the NCgl2816-lldD operon resulted in the inability to grow on l-lactate as a sole carbon source unless the mutant was complemented with a plasmid carrying lldD, indicating that NCgl2816 is not essential for growth on l-lactate. Thus, if NCgl2816 codes for an l-lactate transporter, as suggested from its primary sequence and the operon organization, additional carriers for uptake of l-lactate are likely to exist in C. glutamicum. In fact, l-lactate is imported into E. coli cells either by the l-lactate permease LldP (4) or by the glycolate uptake system GlcA (27). Both systems are redundant with respect to uptake of 2-hydroxymonocarboxylates (26, 27). Since the transcriptome analysis of l-lactate-grown C. glutamicum revealed higher mRNA levels of NCgl2713 and NCgl2965, and both encode putative permeases, these are possible candidates for 2-hydroxymonocarboxylate carriers.
Expression of the NCgl2816-lldD operon was regulated by the carbon source. When l-lactate was present in the medium, increased mRNA levels of NCgl2816 and lldD as well as an increased specific activity of quinone-dependent l-lactate dehydrogenase were observed. Transcription of the lld operon of E. coli is repressed by the two-component signal transduction system ArcB/ArcA during anaerobiosis, is activated by the pyruvate dehydrogenase repressor PdhR, and might be repressed by LldR (4, 32). However, in C. glutamicum, a lactate-responsive regulatory system has not yet been identified.
l-Lactate is a by-product excreted into the medium during glutamate production. In the continuous cultivation of C. glutamicum 2262, the excreted l-lactate was reutilized after the specific activity of quinone-dependent l-lactate dehydrogenase increased; this reutilization might have contributed to glutamate formation. To improve glutamate production, it is proposed to eliminate the concurrent l-lactate formation by deletion of the ldhA gene, since the encoded NAD-dependent l-lactate dehydrogenase was shown to be the only l-lactate-generating enzyme in C. glutamicum (13). Alternatively, increasing the activity of pyruvate dehydrogenase by overexpressing the respective genes or by introducing alleles for deregulated pyruvate dehydrogenase might improve glutamate production, as it was proposed that the reduced activity of pyruvate dehydrogenase observed during glutamate production is responsible for by-product formation (41).
Supplementary Material
Acknowledgments
We are grateful to Jean-Marc Engasser (Nancy, France) and Hermann Sahm (Juelich, Germany) for continuous support.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bott, M., and A. Niebisch. 2003. The respiratory chain of Corynebacterium glutamicum. J. Biotechnol. 104:129-153. [DOI] [PubMed] [Google Scholar]
- 2.Delaunay, S., P. Daran-Lapujade, J. M. Engasser, and J. L. Goergen. 2004. Glutamate as an inhibitor of phosphoenolpyruvate carboxylase activity in Corynebacterium glutamicum. J. Ind. Microbiol. Biotechnol. 31:183-188. [DOI] [PubMed] [Google Scholar]
- 3.Delaunay, S., P. Gourdon, P. Lapujade, E. Mailly, E. Oriol, J. M. Engasser, N. L. Lindley, and J. L. Goergen. 1999. An improved temperature triggered process for glutamate production with Corynebacterium glutamicum. Enzyme Microb. Biotechnol. 25:762-768. [Google Scholar]
- 4.Dong, J. M., J. S. Taylor, D. J. Latour, S. Iuchi, and E. C. Lin. 1993. Three overlapping lct genes involved in l-lactate utilization by Escherichia coli. J. Bacteriol. 175:6671-6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eggeling, L., K. Krumbach, and H. Sahm. 2001. l-Glutamate efflux with Corynebacterium glutamicum: why is penicillin treatment or Tween addition doing the same? J. Mol. Microbiol. Biotechnol. 3:67-68. [PubMed] [Google Scholar]
- 6.Eikmanns, B. J., N. Thum-Schmitz, L. Eggeling, K. U. Lüdtke, and H. Sahm. 1994. Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology 140:1817-1828. [DOI] [PubMed] [Google Scholar]
- 7.Gerstmeir, R., A. Cramer, P. Dangel, S. Schaffer, and B. J. Eikmanns. 2004. RamB, a novel transcriptional regulator of genes involved in acetate metabolism of Corynebacterium glutamicum. J. Bacteriol. 186:2798-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerstmeir, R., V. F. Wendisch, S. Schnicke, H. Ruan, M. Farwick, D. Reinscheid, and B. J. Eikmanns. 2003. Acetate metabolism and its regulation in Corynebacterium glutamicum. J. Biotechnol. 104:99-122. [DOI] [PubMed] [Google Scholar]
- 9.Gutmann, M., C. Hoischen, and R. Kramer. 1992. Carrier-mediated glutamate secretion by Corynebacterium glutamicum under biotin limitation. Biochim. Biophys. Acta 1112:115-123. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 11.Hermann, T. 2003. Industrial production of amino acids by coryneform bacteria. J. Biotechnol. 104:155-172. [DOI] [PubMed] [Google Scholar]
- 12.Hoischen, C., and R. Kramer. 1990. Membrane alteration is necessary but not sufficient for effective glutamate secretion in Corynebacterium glutamicum. J. Bacteriol. 172:3409-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inui, M., S. Murakami, S. Okino, H. Kawaguchi, A. A. Vertes, and H. Yukawa. 2004. Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J. Mol. Microbiol. Biotechnol. 7:182-196. [DOI] [PubMed] [Google Scholar]
- 14.Ishige, T., M. Krause, M. Bott, V. F. Wendisch, and H. Sahm. 2003. The phosphate starvation stimulon of Corynebacterium glutamicum determined by DNA microarray analyses. J. Bacteriol. 185:4519-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalinowski, J., B. Bathe, D. Bartels, N. Bischoff, M. Bott, A. Burkovski, N. Dusch, L. Eggeling, B. J. Eikmanns, L. Gaigalat, A. Goesmann, M. Hartmann, K. Huthmacher, R. Kramer, B. Linke, A. C. McHardy, F. Meyer, B. Mockel, W. Pfefferle, A. Puhler, D. A. Rey, C. Ruckert, O. Rupp, H. Sahm, V. F. Wendisch, I. Wiegrabe, and A. Tauch. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 104:5-25. [DOI] [PubMed] [Google Scholar]
- 16.Keilhauer, C., L. Eggeling, and H. Sahm. 1993. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 175:5595-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura, E., C. Yagoshi, Y. Kawahara, T. Ohsumi, T. Nakamatsu, and H. Tokuda. 1999. Glutamate overproduction in Corynebacterium glutamicum triggered by a decrease in the level of a complex comprising DtsR and a biotin-containing subunit. Biosci. Biotechnol. Biochem. 63:1274-1278. [DOI] [PubMed] [Google Scholar]
- 18.Kinoshita, S., S. Udaka, and M. Shimono. 1957. Studies on amino acid fermentation. Production of l-glutamic acid by various microorganisms. J. Gen. Appl. Microbiol. 3:193-205. [PubMed] [Google Scholar]
- 19.Krug, A., V. F. Wendisch, and M. Bott. 2005. Identification of AcnR, a TetR-type repressor of the aconitase gene acn in Corynebacterium glutamicum. J. Biol. Chem. 280:585-595. [DOI] [PubMed] [Google Scholar]
- 20.Lambert, C., A. Erdmann, M. Eikmanns, and R. Krämer. 1995. Triggering glutamate excretion in Corynebacterium glutamicum by modulating the membrane state with local anesthetics and osmotic gradients. Appl. Environ. Microbiol. 61:4334-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lange, C., D. Rittmann, V. F. Wendisch, M. Bott, and H. Sahm. 2003. Global expression profiling and physiological characterization of Corynebacterium glutamicum grown in the presence of l-valine. Appl. Environ. Microbiol. 69:2521-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marx, A., K. Striegel, A. A. de Graaf, H. Sahm, and L. Eggeling. 1997. Response of the central metabolism of Corynebacterium glutamicum to different flux burdens. Biotechnol. Bioeng. 56:168-180. [DOI] [PubMed] [Google Scholar]
- 23.Molinari, R., and F. J. Lara. 1960. The lactic dehydrogenase of Propionibacterium pentosaceum. Biochem. J. 75:57-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müh, U., V. Massey, and C. H. Williams, Jr. 1994. Lactate monooxygenase. I. Expression of the mycobacterial gene in Escherichia coli and site-directed mutagenesis of lysine 266. J. Biol. Chem. 269:7982-7988. [PubMed] [Google Scholar]
- 25.Nantapong, N., Y. Kugimiya, H. Toyama, O. Adachi, and K. Matsushita. 2004. Effect of NADH dehydrogenase-disruption and over-expression on respiration-related metabolism in Corynebacterium glutamicum KY9714. Appl. Microbiol. Biotechnol. 66:187-193. [DOI] [PubMed] [Google Scholar]
- 26.Nunez, M. F., O. Kwon, T. H. Wilson, J. Aguilar, L. Baldoma, and E. C. Lin. 2002. Transport of l-lactate, d-lactate, and glycolate by the LldP and GlcA membrane carriers of Escherichia coli. Biochem. Biophys. Res. Commun. 290:824-829. [DOI] [PubMed] [Google Scholar]
- 27.Nunez, M. F., M. T. Pellicer, J. Badia, J. Aguilar, and L. Baldoma. 2001. The gene yghK linked to the glc operon of Escherichia coli encodes a permease for glycolate that is structurally and functionally similar to l-lactate permease. Microbiology 147:1069-1077. [DOI] [PubMed] [Google Scholar]
- 27a.Peters-Wendisch, P. G., B. J. Eikmanns, G. Thierbach, B. Bachmann, and H. Sahm. 1993. Phosphoenolpyruvate carboxylase in Corynebacterium glutamicum is dispensable for growth and lysine production. FEMS Microbiol. Lett. 112:269-274. [Google Scholar]
- 28.Peters-Wendisch, P. G., B. Schiel, V. F. Wendisch, E. Katsoulidis, B. Mockel, H. Sahm, and B. J. Eikmanns. 2001. Pyruvate carboxylase is a major bottleneck for glutamate and lysine production by Corynebacterium glutamicum. J. Mol. Microbiol. Biotechnol. 3:295-300. [PubMed] [Google Scholar]
- 29.Polen, T., and V. F. Wendisch. 2004. Genomewide expression analysis in amino acid-producing bacteria using DNA microarrays. Appl. Biochem. Biotechnol. 118:215-232. [DOI] [PubMed] [Google Scholar]
- 30.Pritchard, G. G. 1971. An NAD+-independent l-lactate dehydrogenase from Rhizopus oryzae. Biochim. Biophys. Acta 250:25-34. [DOI] [PubMed] [Google Scholar]
- 31.Puech, V., M. Chami, A. Lemassu, M. A. Laneelle, B. Schiffler, P. Gounon, N. Bayan, R. Benz, and M. Daffe. 2001. Structure of the cell envelope of corynebacteria: importance of the non-covalently bound lipids in the formation of the cell wall permeability barrier and fracture plane. Microbiology 147:1365-1382. [DOI] [PubMed] [Google Scholar]
- 32.Quail, M. A., and J. R. Guest. 1995. Purification, characterization and mode of action of PdhR, the transcriptional repressor of the pdhR-aceEF-lpd operon of Escherichia coli. Mol. Microbiol. 15:519-529. [DOI] [PubMed] [Google Scholar]
- 33.Radmacher, E., K. C. Stansen, G. S. Besra, L. J. Alderwick, W. N. Maughan, G. Hollweg, H. Sahm, V. F. Wendisch, and L. Eggeling. 2005. Ethambutol, a cell wall inhibitor of Mycobacterium tuberculosis, elicits l-glutamate efflux of Corynebacterium glutamicum. Microbiology 151:1359-1368. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 36.Schluesener, D., F. Fischer, J. Kruip, M. Rogner, and A. Poetsch. 2005. Mapping the membrane proteome of Corynebacterium glutamicum. Proteomics 5:1317-1330. [DOI] [PubMed] [Google Scholar]
- 37.Shiio, I., S. I. Ôtsuka, and N. Katsuya. 1963. Cellular permeability and extracellular formation of glutamic acid in Brevibacterium flavum. J. Biochem. 53:333-340. [DOI] [PubMed] [Google Scholar]
- 38.Takinami, K., H. Yoshii, Y. Yamada, H. Okada, and K. Kinoshita. 1968. Control of l-glutamic acid fermentation by biotin and fatty acid. Amino Acid Nucleic Acid Res. 183:120-160. [Google Scholar]
- 39.Tauch, A., O. Kirchner, B. Loffler, S. Gotker, A. Puhler, and J. Kalinowski. 2002. Efficient electrotransformation of Corynebacterium diphtheriae with a mini-replicon derived from the Corynebacterium glutamicum plasmid pGA1. Curr. Microbiol. 45:362-367. [DOI] [PubMed] [Google Scholar]
- 40.Udaka, S. 1960. Screening method for microorganisms accumulating metabolites and its use in the isolation of Micrococcus glutamicus. J. Bacteriol. 79:754-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uy, D., S. Delaunay, P. Germain, J. M. Engasser, and J. L. Goergen. 2003. Instability of glutamate production by Corynebacterium glutamicum 2262 in continuous culture using the temperature-triggered process. J. Biotechnol. 104:173-184. [DOI] [PubMed] [Google Scholar]
- 42.Wendisch, V. F. 2003. Genome-wide expression analysis in Corynebacterium glutamicum using DNA microarrays. J. Biotechnol. 104:273-285. [DOI] [PubMed] [Google Scholar]
- 43.Winnen, B., J. Felce, and M. H. J. Saier. Genomic analyses of transporter proteins in Corynebacterium glutamicum and Corynebacterium efficiens. In L. Eggeling and M. Bott (ed.), Handbook on Corynebacterium glutamicum, in press. CRC Press, Boca Raton, Fla.
- 44.Xia, Z. X., and F. S. Mathews. 1990. Molecular structure of flavocytochrome b2 at 2.4 Å resolution. J. Mol. Biol. 212:837-863. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.