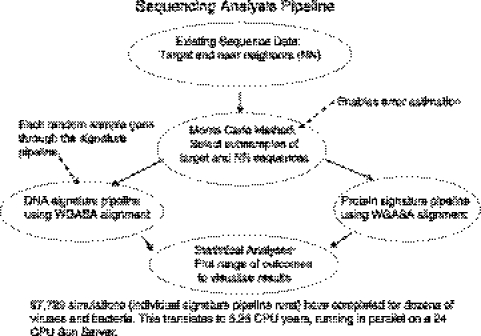
Figure 1.
Diagram of the SAP. For an SAP run, first a pool of target genome and a pool of NN genomes are collected. Then many random subsamples of target and NN genomes are selected from the pool, and each subsample is run through either the DNA signature pipeline or the protein signature pipeline, which identify regions conserved among target genomes and unique relative to non-target genomes, where unique regions are evaluated by comparing to a large sequence database of all currently available bacterial and viral complete genomes or the non-redundant protein database, excluding NNs from the NN pool that are not in that random subsample. Thus, each run of the SAP requires many runs of the DNA or protein signature pipelines with different random samples, generating a range of outcomes that are plotted on range plots.