Figure 5.
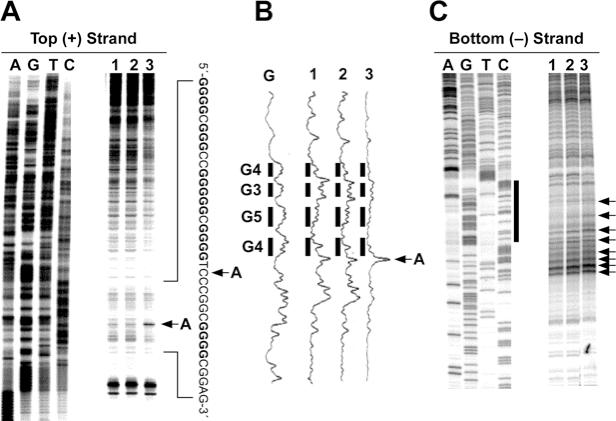
In vitro footprinting of the VEGF promoter region with S1 nuclease. (A) Autoradiograms showing S1 nuclease cleavage sites on the top strand of a supercoiled pGL3-VEGF plasmid. Arrow A indicates the hypersensitive cleavage sites to S1 nuclease. (B) Densitometric scanning of the autoradiogram in (A). The plasmid DNA was incubated in the absence of salt (lane 1) or in the presence of 100 mM KCl without (lane 2) and with (lane 3) 1 µM telomestatin at 37°C for 1 h before digesting with S1 nuclease. S1 nuclease cleavage sites were mapped using linear amplification by PCR with 32P-labeled gene-specific primers on plasmid DNA pretreated with S1 nuclease. Arrow A indicates the hypersensitive cleavage sites to S1 nuclease. (C) Autoradiograms showing S1 cleavage sites on the bottom strand of a supercoiled pGL3-VEGF plasmid. The designation of lanes 1–3 was as in (A) above. The vertical bar next to the gel indicates the polypyrimidine tract and the arrows indicate the S1 nuclease hypersensitivity sites.
