Abstract
Temporal and spatial assembly of signal transduction machinery determines dendrite branch patterning, a process crucial for proper synaptic transmission. Our laboratory previously cloned and characterized cypin, a protein that decreases PSD-95 family member localization and regulates dendrite number. Cypin contains zinc binding, collapsin response mediator protein (CRMP) homology, and PSD-95, Discs large, zona occludens-1 binding domains. Both the zinc binding and CRMP homology domains are needed for dendrite patterning. In addition, cypin binds tubulin via its CRMP homology domain to promote microtubule assembly. Using a yeast two-hybrid screen of a rat brain cDNA library with cypin lacking the carboxyl terminal eight amino acids as bait, we identified snapin as a cypin binding partner. Here, we show by affinity chromatography and coimmunoprecipitation that the carboxyl-terminal coiled-coil domain (H2) of snapin is required for cypin binding. In addition, snapin binds to cypin's CRMP homology domain, which is where tubulin binds. We also show that snapin competes with tubulin for binding to cypin, resulting in decreased microtubule assembly. Subsequently, overexpression of snapin in primary cultures of hippocampal neurons results in decreased primary dendrites present on these neurons and increased probability of branching. Together, our data suggest that snapin regulates dendrite number in developing neurons by modulating cypin-promoted microtubule assembly.
INTRODUCTION
The precise patterning of dendrites is important for determining how information is processed by a neuron (Vetter et al., 2001; Schaefer et al., 2003). When there is an abnormal decrease in the number of dendrite branches on neurons, the neuron cannot receive appropriate information, and hence disruption of proper signaling networks results. Thus, there is great interest in understanding how the number of dendrites produced by a neuron is determined.
A first step for elucidating the mechanism by which dendrite number is regulated is to identify the players in this process. Dendrite arbors are shaped by an interplay between intrinsic and extrinsic factors. Some of the known intrinsic factors are calcium/calmodulin-dependent protein kinase II (Fink et al., 2003); the small GTPases RhoA, Rac1, and Cdc42 (Threadgill et al., 1997; Ruchhoeft et al., 1999; Li et al., 2000); novel genes identified in Drosophila (Gao et al., 1999; Moore et al., 2002; Grueber et al., 2003; Yu and Malenka, 2003; Emoto et al., 2004); β-catenin (Yu and Malenka, 2003); Dishevelled (Rosso et al., 2005); a calcium-responsive transactivator called CREST (Aizawa et al., 2004); and cypin (cytosolic PSD-95 interactor; Firestein et al., 1999; Akum et al., 2004). The external factors comprise a long list and include neurotrophins (McAllister et al., 1995; McAllister et al., 1997; Baker et al., 1998; Horch et al., 1999; Lom and Cohen-Cory, 1999), electrical activity (McAllister et al., 1996; Cambiasso et al., 2000; Vaillant et al., 2002; Yu and Malenka, 2003), and estrogen (Cambiasso et al., 2000; Audesirk et al., 2003a,b; Sakamoto et al., 2003; Dominguez et al., 2004; Nathan et al., 2004). As of yet, there have been only a small number of studies that examine how the extracellular and intrinsic factors are linked to determine dendrite morphology. We have chosen to study the intracellular protein cypin to begin to understand the interplay between extracellular and intrinsic factors because cypin protein levels are increased in response to extracellular factors, such as KCl and nerve growth factor, that increase dendrite number (Akum et al., 2004).
Recently, we reported that cypin acts to increase dendrite number by binding to tubulin heterodimers and by promoting microtubule assembly (Akum et al., 2004). The collapsin response mediator protein (CRMP) homology domain is responsible for this activity (Akum et al., 2004). Because we are interested in elucidating pathways by which dendrite number is regulated, we sought to identify proteins that interact with cypin and that may act to regulate dendrite number as part of a cypin protein complex. Because we have already reported that the carboxy terminus of cypin interacts with PDZ (PSD-95, Discs large, zona occludens-1) domains of members of the PSD-95 family (Firestein et al., 1999), we decided to screen a rat brain library using the yeast two-hybrid system with a cypin mutant lacking the last eight amino acids. We identified snapin, a 15-kDa protein first isolated as a SNAP-25 interacting protein (Ilardi et al., 1999), as a cypin-binding partner.
We further analyzed the interaction between cypin and snapin. We show that snapin and cypin are coimmunoprecipitated from rat brain extracts. Furthermore, using glutathione S-transferase (GST) affinity chromatography, we show that the carboxyl-terminal coiled-coil domain of snapin binds to the CRMP homology domain of cypin and that it competes with tubulin heterodimer binding to cypin. As a result of this competition, we find that the binding of snapin to cypin results in decreased microtubule assembly. In parallel, overexpression of snapin in cultured hippocampal neurons results in decreased primary dendrite number and increased probability of branching. Thus, we have identified snapin as a regulator of dendrite patterning, potentially by modulating cypin-promoted microtubule assembly.
MATERIALS AND METHODS
Antibodies
Rabbit anti-snapin antibodies (against C-terminal peptide and full-length snapin) were purchased from Synaptic Systems (Goettingen, Germany), and monoclonal antibodies recognizing MAP2, synaptophysin, actin, and tubulin were purchased from Sigma-Aldrich (St. Louis, MO). Tetramethylrhodamine B isothiocyanate-labeled phalloidin was also from Sigma-Aldrich. Mouse anti-early endosome antigen (EEA)1 and mouse anti-SNAP-25 were purchased from BD Biosciences (Franklin Lakes, NJ), and mouse anti-TGN 38 and mouse anti-PSD-95 were purchased from Affinity Bioreagents (Golden, CO). Rabbit anti-cypin (RTG-55) and preimmune serum were characterized previously (Akum et al., 2004). Rabbit IgG was purchased from Rockland (Gilbertsville, PA), and Cy2- and Cy3-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). Rabbit anti-GluR1 was purchased from Chemicon International (Temecula, CA). Rat anti-green fluorescent protein (GFP) was a gift from Dr. Shu-Chan Hsu (Rutgers University, Piscataway, NJ).
Yeast Two-Hybrid Screen
cDNA encoding cypin lacking the last eight amino acids was subcloned into pGBKT7 to generate a GAL4-binding domain fusion. This construct was used to screen a library of rat brain cDNAs (BD Biosciences Clontech, Mountain View, CA). β-Galactosidase activity was measured by a colorimetric filter assay. DNA was isolated from colonies that were positive for interaction only with cypin and not with the unrelated lamin C and was sequenced.
Affinity Chromatography
COS-7 cells were transfected with either pEGFP-C1-snapin, pDsRed-N1-snapin, pEGFP-C1-cypin, or pDsRed-N1-cypin using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) as described by the manufacturer. Cells were washed with cold phosphate-buffered saline (PBS) and 1 mM EDTA and scraped into 5 ml of TEEN (25 mM Tris-HCl, pH 7.4, 1 mM EDTA, 1 mM EGTA, 100 mM NaCl). Cells were homogenized using a Potter-Elvehjem tissue grinder (20 strokes). Phenylmethylsulfonyl fluoride (PMSF) and dithiothreitol were added to lysates to final concentrations of 1 mM, and cells were further lysed by passing the extract through a 25-gauge needle 5 times. Lysates were centrifuged at 12,000 × g for 10 min at 4°C. Triton X-100 was added to the supernatant to a final concentration of 1% and incubated at 4°C for 30 min. Lysates were centrifuged at 12,000 × g for 10 min at 4°C. COS-7 cell lysates were incubated with glutathione-Sepharose beads bound to 25 μg of the appropriate GST-fusion proteins for 1 h at 4°C. Beads were washed three times with TEEN. Bound proteins were eluted with 0.5% SDS and 100 mM NaCl. Proteins were resolved on a 10% SDS polyacrylamide gel and transferred to polyvinylidene difluoride (PVDF) membrane. Blots were probed with the indicated antibodies. For experiments using rat brain extracts, the extracts were prepared exactly as described above except that they were not passed through a 25-gauge needle. Experiments were performed in duplicate or triplicate.
Coimmunoprecipitation
For coimmunoprecipitation studies, one rat brain was homogenized in 10 ml of TEE (25 mM Tris, 1 mM EGTA, 1 mM EDTA) + 1 mM PMSF. Triton X-100 was added to a final concentration of 1%, and proteins were extracted for 1 h at 4°C. The extract was centrifuged at 12,000 × g to remove insoluble material, and the supernatant was incubated with either anti-cypin, anti-snapin, preimmune serum, rabbit IgG, anti-tubulin, or mouse IgG. Protein A beads were added and after a 1-h incubation, the beads were washed with TEE + 0.2% Triton X-100. Immunoprecipitated proteins were eluted with protein loading buffer and resolved by SDS-PAGE and transferred to PVDF membrane. Blots were probed with the indicated antibodies. Experiments were performed in duplicate or triplicate.
For quantitation of coimmunoprecipitates, immunoreactive bands were selected from scanned blots, and intensities were quantitated using Adobe Photoshop software. An area close to the bands was used as a reference for background intensity. Number of pixels for the precipitate bands was compared with that of the input (load) to give percentage of precipitated bands. This percentage was then adjusted for amount of input relative to the amount of eluate run for analysis.
Developmental Western Blot
Hippocampal neurons were plated at 1 million cells per 35-mm dishes. At 10, 12, 17, and 24 days in vitro (d.i.v.), cells were washed with ice-cold 1× PBS and scraped into TEE containing 150 mM NaCl and 1 mM PMSF. Cells were homogenized using a Potter-Elvehjem tissue grinder (20 strokes) and further lysed by passing the extract through a 25-gauge needle five times. Lysates were centrifuged at 14,000 × g for 10 min at 4°C. Proteins were resolved on a 15% SDS polyacrylamide gel and transferred to PVDF membrane. The blot was probed with the indicated antibodies.
Synaptosomal Fractionation
Four rat cortices were homogenized in 36 ml of homogenization buffer (320 mM sucrose, 4 mM HEPES, pH 7.4, 1 mM EGTA, 1 mM PMSF) using 10 strokes at 900 rpm of a loose fitting glass-Teflon homogenizer (size 22; Kontes Glass, Vineland, NJ). The homogenate was centrifuged at 1000 × g for 10 min. The supernatant (S1) was collected and centrifuged at 12,000 × g for 15 min, and the pellet (P2) was resuspended in 24 ml of homogenization buffer and centrifuged at 13,000 × g for 15 min. The resulting pellet (P2′), representing a crude synaptosomal fraction, was lysed by osmotic shock and homogenized by three strokes of the glass-Teflon homogenizer at 2000 rpm, and the homogenate was spun at 33,000 × g for 20 min to yield supernatant (LS1) and pellet (LP1, heavy membranes). LS1 was spun at 251,000 × g max for 2 h. The resulting supernatant (LS2) contained soluble proteins, and the pellet (LP2) contained synaptic vesicle proteins. Proteins were resolved on a 10% SDS-polyacrylamide gel, and Western blotting was performed as described above.
Pixel intensities of homogenate, P2, and S2 were determined by using Adobe Photoshop 5. Luminosity of each band was determined using the histogram function. Mean luminosity and total pixels were determined for each of the three samples on each film. Background for each band was taken of an area on the film close to the band. And mean luminosity and total pixels also were determined for background. Mean luminosities were multiplied by total pixels to obtain total luminosity for each band and background. Total luminosity for each band was divided by luminosity of the corresponding background to normalize. Averages were taken for each sample (n = 2), and SEM was determined.
Tubulin Binding Assays
GST, GST-cypin, and GST-snapin were expressed in Escherichia coli and purified using glutathione-Sepharose as described previously (Firestein et al., 1999). Purified proteins were eluted from the beads using glutathione and dialyzed against PBS. Purified cypin (2 μM) and the indicated concentrations of purified snapin were mixed with tubulin heterodimers (7 μM) in PEM buffer (80 mM PIPES, pH 6.9, 2 mM MgCl2, 0.5 mM EGTA) containing 5% glycerol for 4–6 h at 4°C. The mixtures were subjected to immunoprecipitation using a monoclonal antibody raised against tubulin. Binding of purified proteins was assayed by SDS-PAGE followed by Western blotting using a rabbit polyclonal antibody raised against cypin (Akum et al., 2004).
Microtubule Polymerization Assays
Tubulin (30 μM) was mixed with purified protein (2 μM) in PEM buffer containing 5% glycerol and 1 mM GTP on ice. The mixture was then incubated at 37°C, and tubulin polymerization was detected by measuring the absorbance of the solution at 340 nm over time.
Neuronal Culture, Immunohistochemistry, and Transfection
Neuronal cultures were prepared from hippocampi of rat embryos at 18 d gestation. The hippocampi were dissociated by brief mechanical trituration. Cells were plated on poly-d-lysine-coated glass coverslips (12 mm in diameter) at a density of ∼1800 cells/mm2. Cultures were plated and maintained in Neurobasal media supplemented with B27, penicillin, streptomycin, and l-glutamine. For immunocytochemistry, neurons were fixed in 4% paraformaldehyde in PBS for 15 min and labeled with the appropriate antibody. Labeled cells were visualized by immunofluorescence (Olympus IX50 microscope with a Cooke Sensicam charge-coupled device cooled camera, fluorescence, imaging system, and Image Prosoftware). For transfection, neurons were grown for 10 d in culture and transfected with the appropriate constructs using Effectene (QIAGEN, Valencia, CA). Neurons were allowed to express the transfected protein for 48 h and then used for assay of dendrite number.
For immunostaining, neurons were fixed in 4% paraformaldehyde in PBS for 15 min. Cells were then incubated in blocking solution (PBS containing 0.1% Triton X, 2% normal goat serum, 0.02% sodium azide) for 1 h. All antibodies used were diluted in blocking solution. For snapin and TGN staining, 1:500 dilutions of primary antibodies were used. For snapin and synaptophysin staining, 1:250 dilutions of primary antibodies were used. For snapin and early endosome staining, 1:250 dilutions of primary antibodies were used. For snapin-GFP and cypin staining, dilutions of 1:100 for rabbit anti-cypin and 1:1000 for rat anti-GFP were used. All incubations with primary antibodies were performed at room temperature on an orbital shaker for 2 h. Coverslips were then washed with PBS three times. Secondary antibody consisted of a 1:250 dilution of Cy2-conjugated donkey anti-rabbit IgG and Cy3-conjugated donkey anti-mouse IgG. For snapin-GFP and cypin immunostaining, the secondary antibodies were a 1:250 dilution of Cy2-conjugated donkey anti-rat IgG and Cy3-conjugated donkey anti-rabbit IgG. All labeling with secondary antibodies was performed at room temperature on an orbital shaker for 1 h. Washes were performed as stated above. Coverslips were then mounted onto frosted glass microscope slides using Fluormount G.
For confocal analysis, images were collected through a Nikon C1 laser-scanning confocal unit mounted on a Nikon TE300 inverted microscope. A Nikon 60× Plan Apo objective with numerical aperture of 1.4 was used for microscopy. The C1 confocal unit consists of two lasers (Ar and HeNe) for fluorescent excitation at 488 nm and 543 nm and two individual photomultiplier tubes (PTMs) for collecting both channels of fluorescence. Scanning was performed at the resolution of 512 × 512 with the pinhole size set at medium and the PMT gain set at 6.0. Quantitation of snapin immunostaining was performed with Image Pro software.
For intensity studies, hippocampal neurons were stained with snapin primary antibody. Intensities for confocal z-stack images were measured using Image Pro software. Somas for each neuron were traced, and intensities were measured as average pixel intensity within the selected region. Dendrite intensities were measured by tracing dendrites, and intensities were measured as average pixel intensity within selected region.
Guanine Deaminase Activity Assay
COS-7 cells were transfected with pEGFP-C1-snapin, pDsRed-N1-snapin, or pEGFP-C1-cypin using Lipofectamine 2000. Cells were washed once with PBS and scraped into 25 mM Tris-HCl, pH 7.4, and 150 mM NaCl. PMSF was added to a final concentration of 1 mM. The cell suspension was homogenized using a Potter-Elvehjem tissue grinder (20 times). Lysates were then passed through a 25-gauge needle five times and centrifuged at 14,000 rpm at 4°C for 15 min. Then, 50 μl of cypin-GFP lysate was used for each sample. Increasing amounts of either snapin-GFP (N-terminal tagged) or snapin-dsRed (C-terminal tagged) lysates were added to each sample (0, 80, 120, or 200 μl). The remaining volume (to total of 250-μl volume of cell lysate) was lysate from untransfected COS-7 cells. Samples were incubated with 900 μl of either the negative control solution (1 mM 2,4,6-tribromo-3-hyroxybenzoic acid, 0.1 mM 4-amino-antipyrene, 0.025 U/ml xanthine oxidase, 0.00325 U/ml uricase, 0.002U/ml peroxidase in 25 mM Tris-HCl, pH 7.4, 150 mM NaCl) or the assay solution (which contained all the aforementioned ingredients, including 0.5 mM guanine) at 37°C for 15 min. Samples were centrifuged at 12,000 × g for 1 min, and the optical density of each sample was measured at 512 nm after indicated time points.
Assessment of Dendrite Number. Neurons were fixed and stained as described above. Pictures of the transfected neurons were taken as described above. Primary and secondary dendrites were counted as described previously (Akum et al., 2004). The person analyzing the dendrite counts was blinded to the transfection condition. Dendrite counts were performed by at least two people, and a third person unblinded the data. Dendrites were counted if they were ≥3 μm in length (Yu and Malenka, 2003).
Description of Statistics Used to Model Branching Patterns. To model the branching pattern differences between the various constructs, we used Poisson and Binomial generalized linear models (GLM; McCullagh and Nelder, 1999). The number of primary dendrites is assumed to come from a Poisson distribution with a mean primary dendrites number that varies from construct to construct. We attempted to simplify the model by restricting the mean to be equal for a subset of constructs, i.e., group A [GFP and GFP-snapin(1-68)-GFP] and group B [GFP-snapin, GFP-snapin(69-end), and GFP-snapin(81-126)]. We performed an all-subset selection, examining all possible combinations of constructs for which we can estimate a common mean. The final model was selected via minimum Bayesian Information Criterion (BIC) (Schwarz, 1978). We checked the goodness of fit of the Poisson model (p = 0.99) and saw no clear violations of this model assumption.
To examine the proportion of primary dendrites that branch, we used a Binomial generalized linear model. We checked the goodness of fit of the Binomial model (chi-square goodness of fit test, p = 0.15) and saw no consistently clear deviations from the assumed model. Following standard practice of Binomial GLMs, we related the branching proportion to the log of number of primary dendrites via a logit transform. We performed an all-subset selection, examining all possible combinations of constructs for which we can estimate a set of parameters in the Binomial model. The best model, selected by BIC, is one that states that only two distinct distributions are needed: group A [GFP and snapin(1-68)] and group B [snapin(69-end) and snapin(81-126)]. Thus, the snapin, snapin(69-end), and snapin(81-126) constructs lead to an increase in branching proportion that cannot be explained by the decrease in primaries alone. We also analyze the number of secondary dendrites that stem from each primary. We detected no significant differences between any of the constructs. Almost all branching events are bifurcations.
RESULTS
Snapin Is a Binding Partner for Cypin
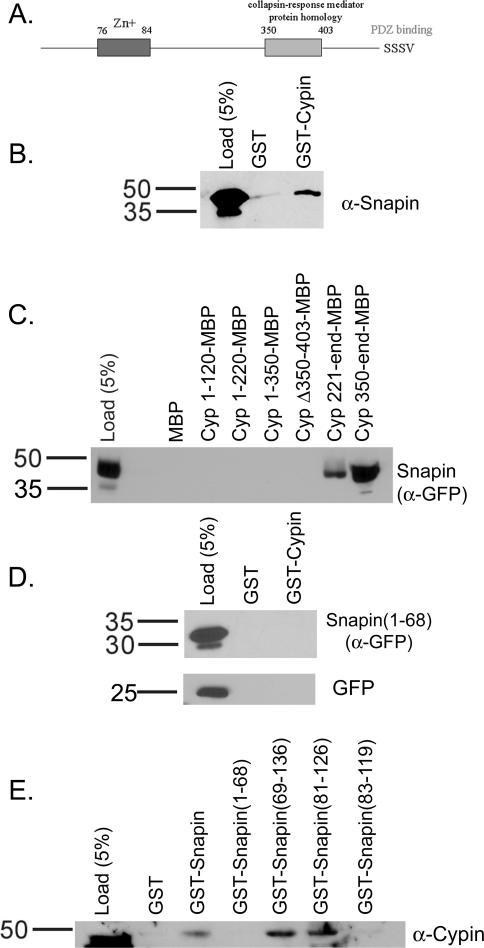
To identify non-PDZ-containing proteins that bind to cypin and may play a role in regulating dendrite number, we screened a rat brain yeast two-hybrid library using cypin lacking the last eight amino acids as bait. We identified 10 potential binding partners, including snapin and the zinc transporter ZnT-3. The cDNAs isolated included the entire coding region for each protein. To identify whether the interaction between cypin and either of these two potential binding partners can occur, we performed GST affinity chromatography using GST-cypin and extracts from rat brain. As shown in Figure 1B, snapin binds to GST-cypin but not GST, indicating that snapin and cypin can interact. We also performed affinity chromatography using extracts from COS-7 cells expressing ZnT-3 or snapin tagged with GFP (Figure 1C; our unpublished data). We detected an interaction between the tagged snapin and cypin (Figure 1C), but we could not detect an interaction between cypin and ZnT-3 (our unpublished data). In addition, we were unable to detect an interaction between cypin and ZnT-3 by affinity chromatography using GST fused to intracellular regions of ZnT-3 or by coimmunoprecipitation from transfected COS-7 cells or from brain extract (our unpublished data). To address whether the interaction between GFP-snapin and cypin is specific, we performed GST-affinity chromatography using extracts from COS-7 cells expressing GFP alone. We found that the GFP itself does not bind to GST-cypin (Figure 1D). These experiments show that snapin is indeed a specific interactor of cypin. Thus, our studies focused on the interaction between cypin and snapin.
Figure 1.
The carboxyl terminal coiled-coil domain of snapin is required to bind to the CRMP homology region of cypin. (A) Schematic of cypin. (B) Detergent soluble extract of rat brain was incubated with glutathione-Sepharose bound to 25 μg of GST or GST-cypin. The Sepharose was washed and eluted, and proteins were resolved by 10% SDS-polyacrylamide electrophoresis and transferred to Immobilon-P. Western blotting of eluates demonstrates that snapin binds to cypin above background binding to GST. Similar results were found when the experiment was performed using COS-7 cells expressing snapin tagged with GFP at its carboxy terminus. (C) Amylose resin bound to 25 μg of maltose binding protein (MBP) fusions of the indicated regions of cypin were incubated with extracts from COS-7 cells expressing snapin tagged with GFP at its amino terminus. Western blotting reveals that snapin binds to cypin when the CRMP homology domain is present (221-end, 350-end) and not when it is absent (1-120, 1-220, 1-350) or deleted (Δ350-403). (D) COS-7 cells were transfected with cDNAs encoding amino acids 1–68 of snapin fused to GFP or GFP alone. Detergent-soluble extracts of these cells were incubated with glutathione-Sepharose bound to 25 μg of GST or GST-Cypin. The Sepharose was washed and eluted, and proteins were resolved by 10% SDS-polyacrylamide electrophoresis and transferred to Immobilon-P. Western blotting of eluates demonstrates that neither snapin(1-68) nor GFP binds to cypin. (E) Extracts from rat brain were incubated with GST fusions of full-length snapin, the first (1-68) and second (69-136) halves of snapin, and the carboxyl-terminal coiled-coil domain (H2) of snapin (defined as either 81-126 or 83-119). The data demonstrate that amino acids 81–126 represent the minimal binding domain of snapin that binds to cypin. Load noted represents the percentage of input material corresponding to the appropriate affinity chromatography.
To identify where snapin binds on cypin, we performed affinity chromatography using maltose binding protein fusions of cypin regions and extracts of COS-7 cells expressing GFP-snapin. As shown in Figure 1C, snapin binds to the C-terminal half of cypin, as evidenced by lack of binding to amino acids 1–120, 1–220, and 1–350 and positive binding to amino acids 221-end and 350-end of cypin. Snapin binds to the region of cypin containing the CRMP homology domain (amino acids 350–403; Figure 1A), as evidenced by binding to amino acids 350-end. Snapin binding was disrupted when the CRMP homology domain was deleted (cypinΔ350-403; Figure 1A). Our data thus suggest that cypin's CRMP homology domain serves as the snapin binding site.
To identify the region of snapin that binds to cypin, we performed GST-affinity chromatography using either the entire 136 amino acids or the first 68 amino acids of snapin on beads and rat brain extract. As shown in Figure 1, D and E, we find that full length but not the first half of snapin binds to cypin. We then determined the region of snapin responsible for cypin binding. Previously, two groups reported the presence of a carboxyl-terminal coiled-coil domain in the second half of snapin (Ilardi et al., 1999; Ruder et al., 2005). This domain was defined as slightly different regions by both groups. To determine which region binds to cypin, we performed GST-affinity chromatography with amino acids 81–126 and 83–119. Surprisingly, only 81–126 bound, suggesting that this is the minimal domain necessary for snapin's binding to cypin. Thus, we find that the carboxyl-terminal coiled-coil domain of snapin is required for interaction with the CRMP homology domain of cypin.
Snapin and Cypin Interact In Vivo
Our data suggest that snapin and cypin could interact in brain. To demonstrate that this is in fact the case, we performed coimmunoprecipitation studies using brain extracts. As shown in Figure 2A, ∼10% of cypin coimmunoprecipitates when all of snapin is precipitated. Furthermore, neither PSD-95 nor SNAP-25 is detected in these immunoprecipitates, showing that the interaction between cypin and snapin is specific. In parallel, ∼20% of snapin coimmunoprecipitates with cypin (Figure 2B). Snapin is not coimmunoprecipitated with tubulin or mouse or rabbit IgG. In addition, a doublet is seen in the cypin immunoprecipitate. The smaller protein may be a degradation product or a posttranslationally modified snapin. As expected, cypin is precipitated when antisera to cypin was used, and tubulin is precipitated when antisera to tubulin is used. Thus, our data suggest that snapin and cypin exist in a complex in the brain.
Figure 2.
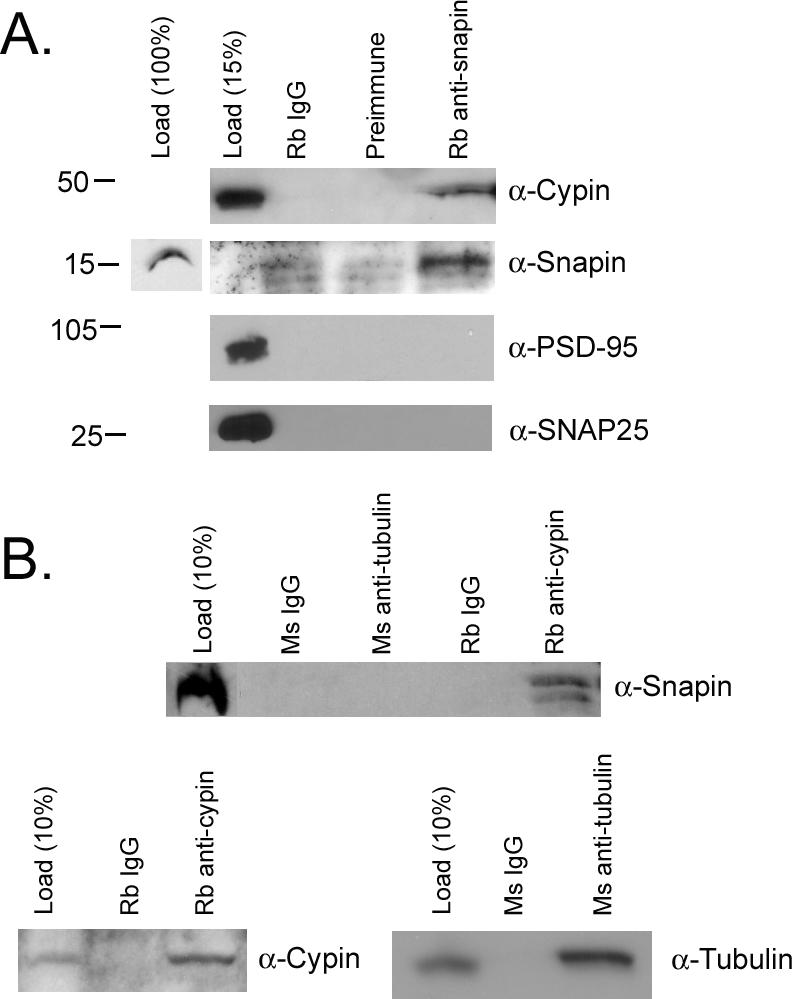
Snapin coimmunoprecipitates with cypin. (A) Detergent-soluble brain extract was incubated with rabbit preimmune serum, rabbit IgG, or rabbit anti-snapin. Antibody complexes were collected with protein A-Sepharose, and the immunoprecipitated complexes were separated on a 10% SDS-polyacrylamide electrophoresis. Western blotting reveals that cypin, but not PSD-95 or SNAP-25, coimmunoprecipitates with snapin. (B) Snapin coimmunoprecipitates with cypin but not tubulin. Western blotting for snapin reveals a doublet at ∼15 and 18 kDa. The 18-kDa form is seen in load. The smaller form found in the immunoprecipitate may represent a degradation product recognized by the antibody. Neither form is seen in precipitates of rabbit or mouse IgG or mouse tubulin. Cypin is precipitated with the cypin antibody, and tubulin is precipitated with the tubulin antibody. Load noted represents the percentage of input material corresponding to the appropriate immunoprecipitation.
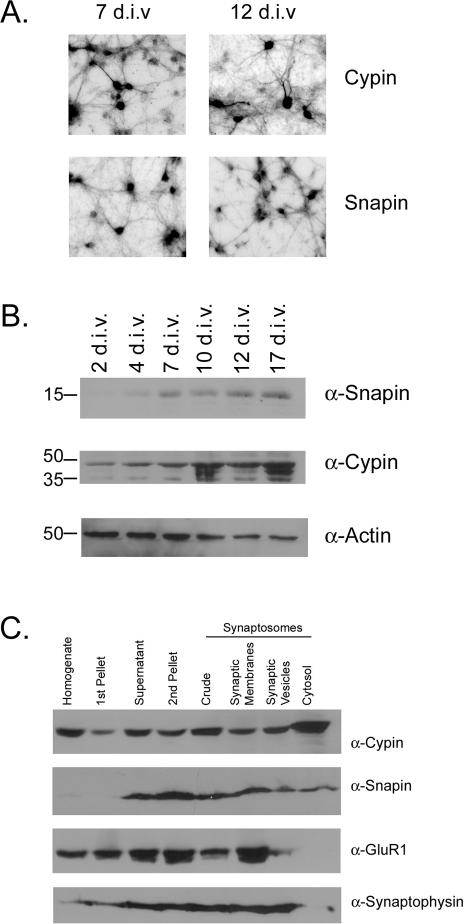
Snapin Is Present in the Cell Body and Dendrites of Developing Neurons
We wanted to assess where snapin is localized in developing neurons to identify what role snapin may play by binding to cypin. We performed immunocytochemistry using cultures of hippocampal neurons at 7 and 12 d.i.v. These time points were selected because cypin plays a role in primary and secondary dendrite development at this time (Akum et al., 2004). As seen in Figure 3A, snapin is found in the cell body and in dendrites. This localization is seen at both time points and is similar to cypin localization. Because both antibodies used for immunostaining were raised in rabbits, we were unable to perform double-staining to compare the endogenous expression patterns of snapin and cypin in a single neuron. As an alternative, we transfected hippocampal neurons with a cDNA encoding GFP-snapin, immunostained for endogenous cypin, and performed confocal imaging. We found that snapin and cypin proteins are both present outside of the nucleus, where they may colocalize (our unpublished data). In addition, there does not seem to be an enrichment of colocalization at any specific organelle (our unpublished data). Thus, our data suggest that snapin and cypin have similar expression patterns in developing hippocampal neurons.
Figure 3.
Snapin is enriched in the cell bodies of developing hippocampal neurons and is found in all synaptosomal fractions. (A) Cultures of primary hippocampal neurons were grown for 7 and 12 d.i.v. and immunostained for snapin or cypin. (B) Snapin, cypin, and actin protein expression were assayed in 15 μg of extracts from neuronal cultures at different developmental time points by Western blotting. Snapin protein expression is low at 2 and 4 d.i.v. when primary dendrites are forming and branching, and expression increases by 7 d.i.v. when primary dendrites have stopped forming and higher order branches are forming. Snapin protein expression remains through 17 d.i.v., when spine formation begins. Cypin protein is expressed at all time points assessed, consistent with the idea that cypin plays a role in dendrite formation and branching. Actin expression serves as a loading control. (C) Snapin is found in all synaptosomal fractions, including synaptic plasma membranes (which are enriched in GluR1), the synaptic vesicle fraction (which is enriched in synaptophysin), and the synaptic cytosol. Like snapin, cypin is enriched in all synaptosomal fractions. Ten micrograms of each fraction was loaded.
To understand at what time during development snapin may be functionally active, we performed Western blotting for snapin protein expression (Figure 3B) and found that snapin protein is expressed at very low levels at 2 d.i.v., when primary branching occurs. Snapin expression increases by 7 d.i.v., when primary dendrite formation is slowing down and higher order branching is occurring. Furthermore, snapin protein expression is maintained by 17 d.i.v. when spine formation is occurring. Thus, the expression pattern of snapin suggests that it may differentially regulate dendrite formation and/or branching.
Snapin and Cypin Are Expressed in a Number of Synaptosomal Fractions
Our data suggest a role for snapin in the cell body or proximal dendrites. However, it is unclear from the literature whether snapin is exclusively localized to synaptic vesicles (Ilardi et al., 1999) or not (Vites et al., 2004). To assess where snapin is localized in rat brain, we performed Western blotting of synaptosomal fractions. Snapin is found in all synaptosomal fractions (Figure 3C). Consistent with our previous report (Firestein et al., 1999), cypin is also found in all synaptosomal fractions. The efficiency of fractionation was verified with GluR1, a membrane receptor subunit that is enriched in the synaptic membrane fraction, and synaptophysin, a synaptic vesicle marker that is enriched in the synaptic vesicle fraction. Our data suggest that snapin may play a role at sites other than synaptic vesicles and that it may regulate cypin function at various sites in the neuron.
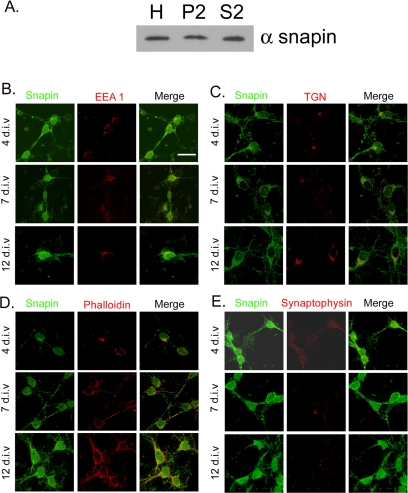
Snapin Is Localized to Both the Membrane and Cytosol
To gain additional insight into how snapin may act to regulate neuronal development, we analyzed the relative amounts of snapin protein in membrane-associated (P2) and soluble (S2) fractions. As seen in Figure 4A, there are equal amounts of snapin in the membrane-associated and soluble fractions (p > 0.05 by t test for band intensity values of 2.38 ± 0.14 [P2] and 2.12 ± 0.54 [S2] arbitrary units over background, n = 2). To determine where in the neuron snapin is localized, we performed confocal analysis of neurons immunostained for snapin and a set of organelle markers. As seen in Figure 4, B, C, and E, snapin does not seem to colocalize with EEA1, an endosomal marker; TGN-38, a trans-Golgi network marker; or synaptophysin, a synaptic marker. A fraction of snapin is found at the plasma membrane (Figure 4D). Thus, snapin seems to be both membrane associated and cytosolic.
Figure 4.
Snapin is membrane associated and cytosolic. (A) Snapin is evenly partitioned between membrane-associated (P2) and soluble fractions (S2). H, unfractionated homogenate. Twenty micrograms of each fraction was loaded. A representative of two different fractionations and Western blots is shown. (B–E) Hippocampal neurons cultured for 4, 7, and 12 d.i.v. were double labeled for snapin and EEA1, an early endosomal marker (B); TGN-38, a trans-Golgi marker (C); phalloidin, a plasma membrane marker (D); or synaptophysin, a synaptic marker (E). The majority of snapin does not colocalize with any of the markers and is absent from the nucleus (E). A fraction of snapin colocalizes with the plasma membrane (D). Bar, 20 μm. The confocal z-sections shown are optimized for organelle localization.
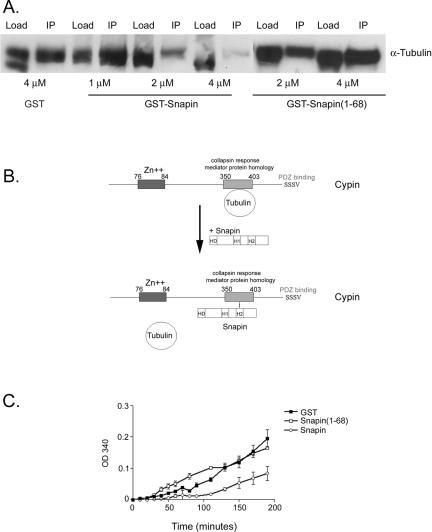
Snapin Competes with Tubulin Heterodimer Binding to Cypin and Slows Cypin-promoted Microtubule Assembly
We next addressed the physiological role of the interaction between snapin and cypin. Because snapin binds to cypin's CRMP homology domain, which is responsible for binding tubulin heterodimers (Akum et al., 2004), we asked whether snapin could compete with tubulin binding to cypin. To address this question, we performed coimmunoprecipitation of purified cypin and tubulin heterodimers in the presence of full-length snapin or the N-terminal half of snapin (amino acids 1–68), which does not bind to cypin. It is important to note that we found no association of snapin with tubulin heterodimers by coimmunoprecipitation or affinity chromatography (our unpublished data). We found that full-length snapin, but not the N-terminal half of snapin or GST, competes with tubulin heterodimer binding to cypin (Figure 5A). In addition, 4 μM snapin almost fully competed with tubulin heterodimer (7 μM) binding to 2 μM cypin. These data suggest a model whereby one tubulin heterodimer binds to the CRMP homology domain of cypin (Akum et al., 2004; Figure 5B). Snapin can compete with this binding, and it has higher affinity for cypin as evidenced by the fact that it takes only 4 μM snapin to compete with 7 μM tubulin from cypin. Furthermore, this competition results in a slower rate of cypin-promoted microtubule assembly that is not seen in the presence of the first half of snapin or GST (Figure 5C). Thus, our data suggest that the binding of snapin to cypin may act to modulate microtubule assembly by regulating tubulin heterodimer binding.
Figure 5.
Snapin competes tubulin heterodimer binding to cypin and slows cypin-promoted microtubule assembly. (A) Purified cypin was mixed with tubulin heterodimers in the presence of increasing amounts of purified snapin and subjected to immunoprecipitation with an antibody raised against cypin. The immunoprecipitates were resolved by SDS-PAGE, and tubulin was detected by Western blotting. Full-length snapin but not the amino terminal half (1-68) competes tubulin binding to cypin. (B) Model for snapin and tubulin binding to cypin. In the absence of snapin (top), one tubulin binds to the CRMP homology domain of cypin. When snapin levels are increased (bottom), snapin competes with tubulin to bind to cypin. Snapin has higher affinity than tubulin for cypin and hence tubulin is released from cypin. (C) Purified snapin proteins were mixed with purified cypin and tubulin heterodimers, and absorbance at 340 nm was taken to assess microtubule polymerization. In the absence of snapin (GST, filled squares), cypin promoted polymerization, whereas the presence of snapin (open circles) slowed down cypin-promoted assembly. The amino terminal half of snapin (1-68, open squares) had no effect on cypin-promoted assembly.
Overexpression of Snapin Results in Changes in Dendrite Branching in Hippocampal Neurons
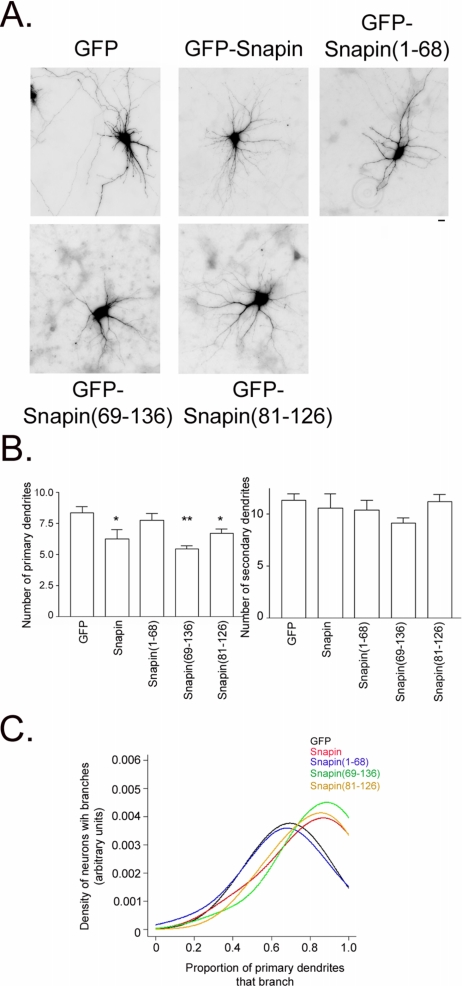
To assess how snapin's modulation of microtubule assembly may affect dendrite morphology, we overexpressed GFP-snapin, GFP-snapin(1-68), GFP-snapin(69-136), GFP-snapin(81-126), or GFP in hippocampal neurons beginning at 10 d.i.v. We then counted primary and secondary dendrites at 12 d.i.v. Overexpression of full-length but not the N-terminal half (1-68) of snapin resulted in decreased primary dendrite number (Figure 6, A and B). In addition, overexpression of the second half (69-136) and the minimal cypin binding region (81-126) also decreased primary dendrite number. Interestingly, none of the snapin proteins had an effect on secondary dendrite number (Figure 6B), thereby increasing the probability of primary dendrite branching (Figure 6C). This means that the number of dendrites that protrude from the cell body (or primary dendrites) is decreased by snapin; however, each of these individual dendrites is more likely to branch, resulting in more secondary dendrites. Compared with neurons expressing GFP alone, neurons overexpressing snapin have a decreased number of primary dendrites, whereas secondary dendrite numbers stay the same. This indicates an increase in the probability of secondary branching when snapin is overexpressed (Figure 6C). To illustrate this point, we graphed the nonparametric estimate of the distribution of the branching probability. The graph clearly indicates that the branching probability is higher for GFP-snapin (red line), GFP-snapin(69-136) (green line), and GFP-snapin(81-126) (yellow line) compared with GFP-snapin(1-68) (blue line) and GFP (black line) as seen in Figure 6C. Furthermore, average dendrite length was unaffected by overexpression of snapin (p = 0.4196). Thus, our data suggest that snapin affects dendrite patterning by decreasing the number of primary dendrites and increasing the probability of dendrite branching.
Figure 6.
Snapin affects dendrite patterning. (A) Representative neurons transfected with cDNA encoding GFP, GFP-snapin, GFP-snapin(1-68), GFP-snapin(69-136), or GFP-snapin(81-126). Bar, 10 μm. (B) Average number of primary and secondary dendrites in neurons that overexpress GFP (n = 74), GFP-snapin (n = 12), GFP-snapin(1-68) (n = 22), GFP-snapin(69-136) (n = 36), or GFP-snapin(81-126) (n = 29). Snapin proteins that bind to cypin [snapin, snapin(69-136), snapin(81-126)] decrease primary dendrite number but do not affect secondary dendrite number. *p < 0.05 and **p < 0.01 by ANOVA followed by Dunnett's multiple comparison test compared with GFP control. (C) Snapin, snapin(69-136), and snapin(81-126) but not snapin(1-68) increase probability of dendrite branching. Data from B were plotted as a distribution of primary dendrites that branch. The graph clearly indicates that the branching probability is higher for snapin (red line), snapin(69-136) (green line), and GFP-snapin(81-126) (yellow line) compared with snapin(1-68) (blue line) and GFP (black line). GFP and GFP-snapin(1-68) do not differ from each other (p > 0.05) nor do GFP-snapin, GFP-snapin(69-136), and GFP-snapin (81-26) differ from each other (p > 0.05), but the two groups differ from each other (p < 0.001).
It is important to note that the GFP, GFP-snapin, and GFP-snapin(81-126) constructs express at similar levels [p > 0.05 by analysis of variance (ANOVA) followed by Bonferroni multiple comparisons test; total fluorescence intensities are as follows: GFP, 3551.14 ± 77.81; GFP-snapin, 3216.15 ± 119.08; and GFP-snapin(81-126), 3351.96 ± 102.05]. GFP-snapin(1-68) and GFP-snapin(69-136) express at lower levels than GFP [p < 0.01; total fluorescence intensities are as follows: GFP-snapin(1-68), 2673.26 ± 164.82 and GFP-snapin(69-136), 2447.71 ± 112.19]; however, this is not an issue because GFP-snapin(69-136) affects branching in a way consistent with the other constructs.
Snapin Is Enriched in the Cell Bodies of Hippocampal Neurons
To address how snapin could differentially affect primary and secondary dendrites, we analyzed a confocal z-stack of neurons immunostained for endogenous snapin (Figure 7A). We analyzed these images for pixel intensities of snapin in the soma versus in the dendrites, and we accounted for the volume difference. Snapin is expressed at a higher level in the cell body than in the dendrites (22.61 ± 0.97 versus 11.15 ± 1.29 average brightness per pixel for five neurons, p < 0.0001 by Student's t test). Because cypin is expressed in both the soma and dendrite (Akum et al., 2004; Figure 3), our data suggest that snapin and cypin may primarily interact in the cell body of developing hippocampal neurons.
Figure 7.
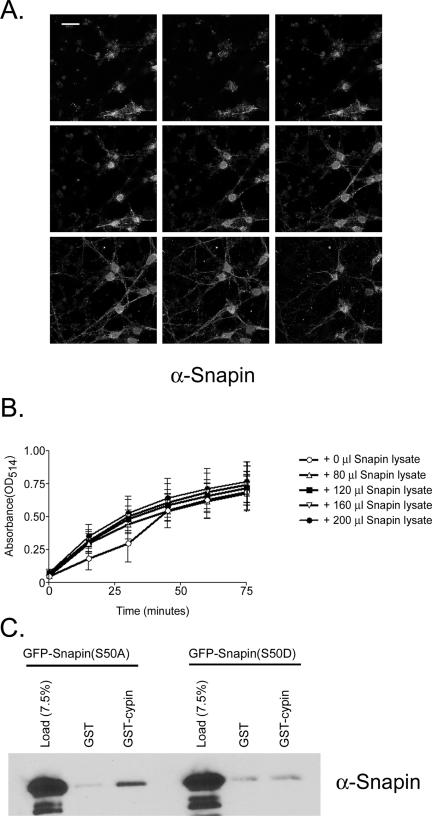
Snapin is enriched in the cell body and does not affect cypin's guanine deaminase activity. (A) Representative confocal z-sections of hippocampal neurons (12 d.i.v.) immunostained for snapin. Z-sections were taken at 1-μm intervals. Bar, 20 μm. (B) Snapin does not influence the guanine deaminase activity of cypin. Lysate (50 μl) from COS-7 cells expressing cypin-GFP was mixed with lysates (0, 80, 120, or 200 μl) of COS-7 cells expressing snapin-GFP (N-terminal tagged) or snapin-DsRed (C-terminal tagged). The remaining volume (to total of 250-μl volume of cell lysate) was lysate from untransfected COS-7 cells. This mixture was then assayed for guanine deaminase activity using a colorometric assay. n = 4 assays, each performed in duplicate. p > 0.05 as determined by ANOVA. (C) Unphosphorylated snapin binds to cypin better than a phosphomimetic form of snapin. Extracts from COS-7 cells expressing either GFP-snapin(S50A) or GFP-snapin(S50D) were incubated with GST or GST-cypin bound to glutathione beads. Eluates were analyzed for the presence of snapin by SDS-PAGE and Western blotting using a polyclonal antibody to snapin. A representative blot is shown for three different experiments.
Snapin Does Not Affect the Guanine Deaminase Activity of Cypin
Because snapin changes dendrite patterning, and cypin's guanine deaminase activity plays a role in regulating dendrite patterning (Akum et al., 2004), we asked whether snapin could regulate cypin's ability to metabolize guanine. We assayed guanine deaminase activity in cellular extracts from COS-7 cells that overexpress snapin and/or cypin. Interestingly, we did not see any effect of snapin on cypin's guanine deaminase activity (Figure 7B). Thus, we think that snapin may regulate dendrite patterning by regulating cypin-promoted microtubule assembly and not by regulating cypin's guanine deaminase activity.
Phosphorylation of Snapin Decreases Binding to Cypin
Phosphorylation of snapin by PKA strengthens its association with SNAP-25 (Chheda et al., 2001). To address whether phosphorylation can play a role in regulating snapin's interaction with cypin, we expressed point mutants of snapin that mimic the constitutively unphosphorylated form (S50A) or the phosphorylated form (S50D). As seen in Figure 7C, only the unphosphorylated form binds to cypin over background levels (i.e., binding to GST). Our data support the idea that phosphorylation of snapin negatively regulates its association with cypin.
DISCUSSION
The primary finding of our study is that snapin, a protein previously thought to play only a presynaptic role in the neuron (Ilardi et al., 1999; Chheda et al., 2001; Evans and Morgan, 2003; Morenilla-Palao et al., 2004; Thakur et al., 2004; Ruder et al., 2005), also plays an important role in regulating dendrite patterning in hippocampal neurons. Similar to data shown by Jahn and colleagues (Vites et al., 2004), we find that snapin is found in all synaptosomal fractions, and we show that it cofractionates and coimmunoprecipitates with cypin. Furthermore, we show that snapin is expressed in the cell bodies and processes of developing hippocampal neurons, suggesting a developmental role for snapin in addition to synaptic vesicle fusion.
Our most exciting finding is that snapin does not merely change the number of dendrites found on a neuron but that it changes the actual pattern of these dendrites. This pattern would be missed by merely counting branch tips as is often done, and thus, counting primary and secondary dendrites is the best method for looking at branching patterns proximal to the cell body. Furthermore, our data suggest that snapin decreases primary dendrites and that it increases the probability of branching. In fact, the best model of probability of branching is one that states that there are two categories into which snapin constructs fall: 1) GFP and GFP-snapin(1-68) with probability of branching ranging from 0.80 (for n = 4 primaries) to 0.65 (for n = 10 primaries); and 2) GFP-snapin, GFP-snapin(69-136) and GFP-snapin(81-126) with branching proportion ranging from 0.85 to 0.75. Thus, the snapin, snapin(69-136) and snapin(81-126) constructs lead to an increase in probability of branching that cannot be explained by the decrease in primaries alone. If the increase in probability of branching by snapin were to be solely due to a decrease in primary dendrites, the range of branching probabilities would be the same for both groups of constructs. However, the difference between the two construct groups is highly significant (p < 0.01). Thus, we think that this is the first report of a protein that differentially affects primary and secondary dendrites. Because the precise patterning of dendrites is important for determining how information is processed by a neuron (Vetter et al., 2001; Schaefer et al., 2003), snapin plays a role in neuronal function by affecting dendrite patterning.
How can snapin play such a role in dendrite patterning? One possibility suggested by our data is that snapin can bind to the CRMP homology region of cypin. We have previously shown that this region binds tubulin heterodimers and mediates microtubule assembly (Akum et al., 2004). A mutant cypin protein that lacks the CRMP homology domain blocks cypin-promoted increases in dendrite number, suggesting that microtubule assembly is important for regulating dendrite number (Akum et al., 2004). We now report that snapin competes with tubulin heterodimer for binding to cypin, resulting in slowed microtubule assembly. In fact, snapin binds cypin with higher affinity than does tubulin. This is physiologically relevant because there is a much higher concentration of tubulin in the neuron than of snapin. Thus, for snapin to have an effect of cypin function, it must compete with this large pool of tubulin.
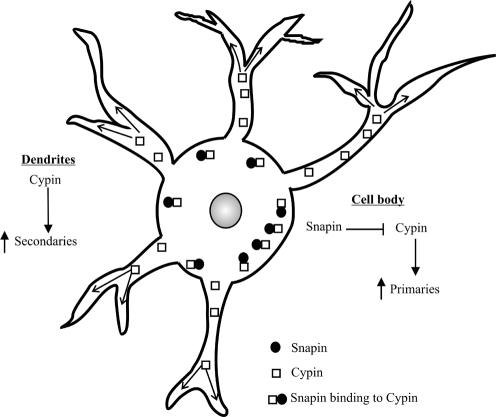
Why are primary and secondary dendrites differentially regulated? We see that snapin protein levels are higher in the cell body than in the dendrites of developing hippocampal neurons (Figure 7). We have previously shown that cypin is expressed throughout the cell body and processes (Firestein et al., 1999; Akum et al., 2004). One mechanism by which snapin may act is by competing tubulin heterodimers from cypin in the cell body, where primary dendrites originate, thereby inhibiting cypin-promoted microtubule assembly locally. This in turn would attenuate primary dendrite formation. We do not know why snapin increases the branching probability of these primary dendrites. There is less snapin in the dendrites than in the cell body; therefore, cypin would be expected to promote microtubule assembly in primary dendrites, leading to secondary dendrite formation. One hypothesis is that by decreasing primary dendrite formation, limiting reagents, perhaps tubulin, may be freed up for secondary dendrite formation. A model for snapin action is demonstrated in Figure 8.
Figure 8.
Model for snapin action. Snapin protein is expressed in the cell bodies of developing neurons where it can interact with cypin and inhibit microtubule assembly. This results in decreased primary dendrite production. Snapin is expressed at lower levels in the dendrites than in the cell bodies, where cypin can promote microtubule assembly in primary dendrites. Hence, branching occurs and secondary dendrites are formed.
It is important to note that cypin is an enzyme involved in guanine metabolism (Yuan et al., 1999). Furthermore, this activity plays a role in regulating dendrite number in hippocampal neurons. Snapin could potentially modulate cypin's enzymatic activity. However, using guanine deaminase assays in extracts from COS-7 cells that overexpress snapin and/or cypin, we did not see effects of snapin on cypin's guanine deaminase activity (Figure 7). Thus, we think that snapin acts to modulate cypin-promoted microtubule assembly but does not affect cypin's enzymatic activity.
We have also found that we cannot coimmunoprecipitate SNAP-25 with snapin from rat brain extracts (Figure 2). However, we are not debating whether a SNAP-25–snapin complex exists, but rather that the amount of this complex may be smaller than a cypin–snapin complex, with 20% of snapin associating with cypin. Furthermore, we found that snapin is expressed in sites in the neuron other than synaptic vesicles (Figure 3), with the protein being both membrane-associated and cytosolic (Figure 4A). Indeed, these results overlap with data published by Rowe and colleagues showing a diffuse cytosolic distribution of snapin (Buxton et al., 2003). In fact, this group also showed that the same region of snapin that binds cypin, the carboxyl-terminal coiled-coil domain (H2), is involved in binding SNAP-23, suggesting that the snapin–SNAP-23 (and snapin:SNAP-25) complex is separate from the snapin–cypin complex. Thus, we think that snapin may have different functions depending on its site in the neuron and on the binding partners with which it interacts.
What signaling pathways may be involved in snapin's regulation of dendrite patterning? One candidate is the receptor tyrosine kinase MET, a receptor for the hepatocyte growth factor/scatter factor (HGF), which has been reported to enhance neurite outgrowth in neocortical explants (Hamanoue et al., 1996) and sympathetic neurons (Maina et al., 1998). MET has been shown to directly interact with snapin (Schaaf et al., 2005), and thus snapin may mediate the effects of this receptor on dendrite patterning. In addition, snapin is phosphorylated by cAMP-dependent protein kinase (PKA; Chheda et al., 2001), and it has been shown that PKA is involved in HGF signaling in kidney (Santos et al., 1993). Interestingly, our data suggest that the unphosphorylated snapin binds better to cypin than a phosphomimetic, supporting the idea that phosphorylation of snapin may favor a snapin–SNAP-25 complex, whereas dephosphorylation of snapin would favor the formation of a snapin–cypin complex. PKA is thought to play a role in dendrite patterning (Audesirk et al., 1997; Chen et al., 2005), and one model for its action would be the regulation of snapin with its binding partners.
Our current study focuses on the role of the snapin–cypin complex in neurons. However, both of these proteins are also found in tissues other than brain. In fact, it has been recently reported that snapin is part of the biogenesis of lysosome-related organelles complex-1 (BLOC-1; Starcevic and Dell'Angelica, 2004). Lysosome-related organelles are cell type-specific organelles found in melanosomes, plateletdense bodies, and cytotoxic T-cell granules (Bonifacino, 2004). Thus, it is possible that the snapin–cypin complex may play a role in cell type-specific functions, for example, in BLOC-1 function, in nonneuronal tissues.
Acknowledgments
We thank Drs. Chris Rongo and Erik Charych and members of the Firestein laboratory for comments on the manuscript, and Dr. James Zheng for help with confocal imaging. B.L.F. thanks Max Miller for inspiration every day. This work was supported in part by a Busch Biomedical Grant, National Science Foundation Grant IBN-0234206, March of Dimes Foundation Grant 1-FY04-107 (to B.L.F), a Rutgers University Undergraduate Research Fellowship (to G. B.), and National Science Foundation Grant DMS-0306360 (to R.J.J.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–02–0165) on August 24, 2005.
Abbreviations used: CRMP, collapsin response mediator protein; cypin, cytosolic PSD-95 interactor; PDZ, PSD-95, Discs large, zona occludens-1; PKA, cAMP-dependent protein kinase; PSD, postsynaptic density; snapin, SNAP-25 interacting protein.
References
- Aizawa, H., Hu, S. C., Bobb, K., Balakrishnan, K., Ince, G., Gurevich, I., Cowan, M., and Ghosh, A. (2004). Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science 303, 197–202. [DOI] [PubMed] [Google Scholar]
- Akum, B. F., Chen, M., Gunderson, S. I., Riefler, G. M., Scerri-Hansen, M. M., and Firestein, B. L. (2004). Cypin regulates dendrite patterning in hippocampal neurons by promoting microtubule assembly. Nat. Neurosci. 7, 145–152. [DOI] [PubMed] [Google Scholar]
- Audesirk, G., Cabell, L., and Kern, M. (1997). Modulation of neurite branching by protein phosphorylation in cultured rat hippocampal neurons. Brain Res. Dev. Brain Res. 102, 247–260. [DOI] [PubMed] [Google Scholar]
- Audesirk, T., Cabell, L., Kern, M., and Audesirk, G. (2003a). beta-estradiol influences differentiation of hippocampal neurons in vitro through an estrogen receptor-mediated process. Neuroscience 121, 927–934. [DOI] [PubMed] [Google Scholar]
- Audesirk, T., Cabell, L., Kern, M., and Audesirk, G. (2003b). Enhancement of dendritic branching in cultured hippocampal neurons by 17beta-estradiol is mediated by nitric oxide. Int. J. Dev. Neurosci. 21, 225–233. [DOI] [PubMed] [Google Scholar]
- Baker, R. E., Dijkhuizen, P. A., Van Pelt, J., and Verhaagen, J. (1998). Growth of pyramidal, but not non-pyramidal, dendrites in long-term organotypic explants of neonatal rat neocortex chronically exposed to neurotrophin-3. Eur. J. Neurosci. 10, 1037–1044. [DOI] [PubMed] [Google Scholar]
- Bonifacino, J. S. (2004). Insights into the biogenesis of lysosome-related organelles from the study of the hermansky-pudlak syndrome. Ann. N.Y. Acad. Sci. 1038, 103–114. [DOI] [PubMed] [Google Scholar]
- Buxton, P., Zhang, X. M., Walsh, B., Sriratana, A., Schenberg, I., Manickam, E., and Rowe, T. (2003). Identification and characterization of Snapin as a ubiquitously expressed SNARE-binding protein that interacts with SNAP23 in non-neuronal cells. Biochem. J. 375, 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambiasso, M. J., Colombo, J. A., and Carrer, H. F. (2000). Differential effect of oestradiol and astroglia-conditioned media on the growth of hypothalamic neurons from male and female rat brains. Eur. J. Neurosci. 12, 2291–2298. [DOI] [PubMed] [Google Scholar]
- Chen, Y., Wang, P. Y., and Ghosh, A. (2005). Regulation of cortical dendrite development by Rap1 signaling. Mol. Cell Neurosci. 28, 215–228. [DOI] [PubMed] [Google Scholar]
- Chheda, M. G., Ashery, U., Thakur, P., Rettig, J., and Sheng, Z. H. (2001). Phosphorylation of Snapin by PKA modulates its interaction with the SNARE complex. Nat. Cell Biol. 3, 331–338. [DOI] [PubMed] [Google Scholar]
- Dominguez, R., Jalali, C., and de Lacalle, S. (2004). Morphological effects of estrogen on cholinergic neurons in vitro involves activation of extracellular signal-regulated kinases. J. Neurosci. 24, 982–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto, K., He, Y., Ye, B., Grueber, W. B., Adler, P. N., Jan, L. Y., and Jan, Y. N. (2004). Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell 119, 245–256. [DOI] [PubMed] [Google Scholar]
- Evans, G. J., and Morgan, A. (2003). Regulation of the exocytotic machinery by cAMP-dependent protein kinase: implications for presynaptic plasticity. Biochem. Soc. Trans. 31, 824–827. [DOI] [PubMed] [Google Scholar]
- Fink, C. C., Bayer, K. U., Myers, J. W., Ferrell, J. E., Jr., Schulman, H., and Meyer, T. (2003). Selective regulation of neurite extension and synapse formation by the beta but not the alpha isoform of CaMKII. Neuron 39, 283–297. [DOI] [PubMed] [Google Scholar]
- Firestein, B. L., Brenman, J. E., Aoki, C., Sanchez-Perez, A. M., El-Husseini, A. E., and Bredt, D. S. (1999). Cypin: a cytosolic regulator of PSD-95 postsynaptic targeting. Neuron 24, 659–672. [DOI] [PubMed] [Google Scholar]
- Gao, F. B., Brenman, J. E., Jan, L. Y., and Jan, Y. N. (1999). Genes regulating dendritic outgrowth, branching, and routing in Drosophila. Genes Dev. 13, 2549–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber, W. B., Jan, L. Y., and Jan, Y. N. (2003). Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell 112, 805–818. [DOI] [PubMed] [Google Scholar]
- Hamanoue, M., Takemoto, N., Matsumoto, K., Nakamura, T., Nakajima, K., and Kohsaka, S. (1996). Neurotrophic effect of hepatocyte growth factor on central nervous system neurons in vitro. J. Neurosci. Res. 43, 554–564. [DOI] [PubMed] [Google Scholar]
- Horch, H. W., Kruttgen, A., Portbury, S. D., and Katz, L. C. (1999). Destabilization of cortical dendrites and spines by BDNF. Neuron 23, 353–364. [DOI] [PubMed] [Google Scholar]
- Ilardi, J. M., Mochida, S., and Sheng, Z. H. (1999). Snapin: a SNARE-associated protein implicated in synaptic transmission. Nat. Neurosci. 2, 119–124. [DOI] [PubMed] [Google Scholar]
- Li, Z., Van Aelst, L., and Cline, H. T. (2000). Rho GTPases regulate distinct aspects of dendritic arbor growth in Xenopus central neurons in vivo. Nat. Neurosci. 3, 217–225. [DOI] [PubMed] [Google Scholar]
- Lom, B., and Cohen-Cory, S. (1999). Brain-derived neurotrophic factor differentially regulates retinal ganglion cell dendritic and axonal arborization in vivo. J. Neurosci. 19, 9928–9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maina, F., Hilton, M. C., Andres, R., Wyatt, S., Klein, R., and Davies, A. M. (1998). Multiple roles for hepatocyte growth factor in sympathetic neuron development. Neuron 20, 835–846. [DOI] [PubMed] [Google Scholar]
- McAllister, A. K., Katz, L. C., and Lo, D. C. (1996). Neurotrophin regulation of cortical dendritic growth requires activity. Neuron 17, 1057–1064. [DOI] [PubMed] [Google Scholar]
- McAllister, A. K., Katz, L. C., and Lo, D. C. (1997). Opposing roles for endogenous BDNF and NT-3 in regulating cortical dendritic growth. Neuron 18, 767–778. [DOI] [PubMed] [Google Scholar]
- McAllister, A. K., Lo, D. C., and Katz, L. C. (1995). Neurotrophins regulate dendritic growth in developing visual cortex. Neuron 15, 791–803. [DOI] [PubMed] [Google Scholar]
- McCullagh, P., and Nelder, J. A. (1999). Generalized Linear Models, Boca Raton, FL: Chapman & Hall/CRC.
- Moore, A. W., Jan, L. Y., and Jan, Y. N. (2002). Hamlet, a binary genetic switch between single- and multiple-dendrite neuron morphology. Science 297, 1355–1358. [DOI] [PubMed] [Google Scholar]
- Morenilla-Palao, C., Planells-Cases, R., Garcia-Sanz, N., and Ferrer-Montiel, A. (2004). Regulated exocytosis contributes to protein kinase C potentiation of vanilloid receptor activity. J. Biol. Chem. 279, 25665–25672. [DOI] [PubMed] [Google Scholar]
- Nathan, B. P., Barsukova, A. G., Shen, F., McAsey, M., and Struble, R. G. (2004). Estrogen facilitates neurite extension via apolipoprotein E in cultured adult mouse cortical neurons. Endocrinology 145, 3065–3073. [DOI] [PubMed] [Google Scholar]
- Rosso, S. B., Sussman, D., Wynshaw-Boris, A., and Salinas, P. C. (2005). Wnt signaling through Dishevelled, Rac and JNK regulates dendritic development. Nat. Neurosci. 8, 34–42. [DOI] [PubMed] [Google Scholar]
- Ruchhoeft, M. L., Ohnuma, S., McNeill, L., Holt, C. E., and Harris, W. A. (1999). The neuronal architecture of Xenopus retinal ganglion cells is sculpted by rho-family GTPases in vivo. J. Neurosci. 19, 8454–8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruder, C., Reimer, T., Delgado-Martinez, I., Hermosilla, R., Engelsberg, A., Nehring, R., Dorken, B., and Rehm, A. (2005). EBAG9 adds a new layer of control on large dense-core vesicle exocytosis via interaction with Snapin. Mol. Biol. Cell 16, 1245–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, H., Mezaki, Y., Shikimi, H., Ukena, K., and Tsutsui, K. (2003). Dendritic growth and spine formation in response to estrogen in the developing Purkinje cell. Endocrinology 144, 4466–4477. [DOI] [PubMed] [Google Scholar]
- Santos, O. F., Moura, L. A., Rosen, E. M., and Nigam, S. K. (1993). Modulation of HGF-induced tubulogenesis and branching by multiple phosphorylation mechanisms. Dev. Biol. 159, 535–548. [DOI] [PubMed] [Google Scholar]
- Schaaf, C. P., Benzing, J., Schmitt, T., Erz, D. H., Tewes, M., Bartram, C. R., and Janssen, J. W. (2005). Novel interaction partners of the TPR/MET tyrosine kinase. FASEB J. 19, 267–269. [DOI] [PubMed] [Google Scholar]
- Schaefer, A. T., Larkum, M. E., Sakmann, B., and Roth, A. (2003). Coincidence detection in pyramidal neurons is tuned by their dendritic branching pattern. J. Neurophysiol. 89, 3143–3154. [DOI] [PubMed] [Google Scholar]
- Schwarz, G. (1978). Estimating the dimension of a model. Ann. Stat. 6, 461–464. [Google Scholar]
- Starcevic, M., and Dell'Angelica, E. C. (2004). Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1). J. Biol. Chem. 279, 28393–28401. [DOI] [PubMed] [Google Scholar]
- Thakur, P., Stevens, D. R., Sheng, Z. H., and Rettig, J. (2004). Effects of PKA-mediated phosphorylation of Snapin on synaptic transmission in cultured hippocampal neurons. J. Neurosci. 24, 6476–6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill, R., Bobb, K., and Ghosh, A. (1997). Regulation of dendritic growth and remodeling by Rho, Rac, and Cdc42. Neuron 19, 625–634. [DOI] [PubMed] [Google Scholar]
- Vaillant, A. R., Zanassi, P., Walsh, G. S., Aumont, A., Alonso, A., and Miller, F. D. (2002). Signaling mechanisms underlying reversible, activity-dependent dendrite formation. Neuron 34, 985–998. [DOI] [PubMed] [Google Scholar]
- Vetter, P., Roth, A., and Hausser, M. (2001). Propagation of action potentials in dendrites depends on dendritic morphology. J. Neurophysiol. 85, 926–937. [DOI] [PubMed] [Google Scholar]
- Vites, O., Rhee, J. S., Schwarz, M., Rosenmund, C., and Jahn, R. (2004). Reinvestigation of the role of snapin in neurotransmitter release. J. Biol. Chem. 279, 26251–26256. [DOI] [PubMed] [Google Scholar]
- Yu, X., and Malenka, R. C. (2003). Beta-catenin is critical for dendritic morphogenesis. Nat. Neurosci. 6, 1169–1177. [DOI] [PubMed] [Google Scholar]
- Yuan, G., Bin, J. C., McKay, D. J., and Snyder, F. F. (1999). Cloning and characterization of human guanine deaminase. Purification and partial amino acid sequence of the mouse protein. J. Biol. Chem. 274, 8175–8180. [DOI] [PubMed] [Google Scholar]