Abstract
A total of 118 fluorescent pseudomonads associated with hazelnut decline, which has been occurring for many years in different areas of northern Greece and Italy, were assessed by performing a repetitive PCR analysis with enterobacterial repetitive intergenic consensus, box element, and repetive extragenic palindromic primer sets, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of whole-cell protein extracts, a carbon compound utilization analysis, and an analysis to determine the presence of the syrB gene. A subset of 53 strains was also characterized by amplified 16S ribosomal DNA restriction analysis (ARDRA) by using nine restriction endonucleases. The virulence of 40 representative strains was assessed by using serial doses. The pathogenic specificities of the strains were also verified. ARDRA carried out with HinfI revealed two main groups of strains, groups A and B, which exhibited a level of similarity of 57%. The other eight restriction endonucleases used did not separate the strains. In addition, a cluster analysis performed by the unweighted pair group method using arithmetic averages after repetitive PCR and SDS-PAGE of protein extracts also revealed the same two groups. Furthermore, the differential utilization of some carbon compounds made it possible to differentiate the groups. Virulence assessment clearly indicated that the group A strains are very virulent, whereas the group B strains proved to be mildly virulent for hazelnut. Group A included the strains isolated in northern Greece and central Italy (i.e., the province of Viterbo); these strains do not have the syrB gene, are pathogenically restricted to Corylus avellana, and belong to Pseudomonas avellanae. Group B includes the other strains obtained from hazelnut cultivated in Piedmont, Campania, Latium, Sicily, and Sardinia. They represent a distinct taxon closely related to Pseudomonas syringae pv. syringae.
Pseudomonas avellanae, the causative agent of bacterial canker and decline of hazelnut (Corylus avellana L.) (5), is severely damaging the cultivated hazelnuts in northern Greece (13) and central Italy (18). In the latter area, where specialized cultivation of hazelnut occurs on more than 20,000 ha, around 1,000 ha is threatened by the bacterium and more than 40,000 trees have already died. The main field symptom is sudden wilting of the twigs, branches, and tree, especially at the end of spring and in the summer. When inoculated in early autumn through leaf scars, the pathogen can systemically reach the roots and then the other branches of a tree (18). Field surveys in the hazelnut-growing areas of Campania, Latium, Piedmont, Sardinia, and Sicily have been performed to ascertain the possible presence and relevance of symptoms resembling those of P. avellanae infection and to isolate the microorganisms responsible for the disease. Previous studies showed that the strains in northern Greece can be differentiated from those in central Italy by repetitive PCR (17). Moreover, we ascertained that in a small district in northwest Italy, where hazelnut is cultivated on around 6,000 ha, there is a composite population of the pathogen that causes wilting of the branches. In this area complete death of a tree was rarely observed, and the populations found proved to be less virulent for C. avellana than the populations isolated in northern Greece and central Italy (19). Genomic fingerprinting of the northwest Italy populations by repetitive PCR performed with (ERIC, BOX, and REP, respectively) primer sets revealed a level of similarity of only 20% with the P. avellanae populations of northern Greece and central Italy (19).
To further investigate the genomic relationships among the pseudomonads that cause wilting of hazelnut trees, we included in our studies a technique that, on the basis of a comparison of conserved regions of the bacterial genome, allowed us to differentiate bacteria at the species or subspecies level. This technique, amplified ribosomal DNA (rDNA) restriction analysis (ARDRA), proved to be very effective for discriminating among Pseudomonas species (1, 7, 12, 14). In parallel, we investigated both the virulence of each of the different populations for hazelnut and the pathogenic specificities of the populations.
In this paper we describe P. avellanae populations that cause localized epidemics and kill thousands of hazelnut trees, as well other pseudomonads that are pathogenic for C. avellana and are genetically related to P. avellanae at a level of 57%; at this time the latter organisms are in an endemic phase and are causing the death of twigs, branches, and, rarely, trees.
MATERIALS AND METHODS
Strains and culture conditions.
Table 1 shows the strains used in this study. All strains were routinely grown on nutrient sucrose agar (NSA) containing 28.0 g of nutrient agar (Oxoid), 5.0 of sucrose, and 1,000 ml of distilled water. All cultures were incubated at 25 to 27°C. Strains were also stored at −20°C in 523 broth medium containing 10.0 g of sucrose, 4.0 g of yeast extract, 8.0 g of casein acid hydrolysate, 2.0 g of K2HPO4, 0.3 g of MgSO4 · 7H2O, and 1,000 ml of distilled water.
TABLE 1.
P. avellanae strains and related pseudomonad strains isolated from C. avellana used in repetitve PCR genomic fingerprinting, whole-cell protein, and carbon compound utilization analyses
| Straina | Region | Year of isolation | Cultivar or species | Straina | Region | Year of isolation | Cultivar or species | |
|---|---|---|---|---|---|---|---|---|
| Strains from Greece | ||||||||
| BPIC 631Tbcd | Drama | 1976 | Palaz | |||||
| BPIC 632c | Drama | 1976 | Palaz | |||||
| BPIC 708c | Drama | 1987 | Palaz | |||||
| BPIC 710 | Drama | 1987 | Palaz | |||||
| BPIC 714cd | Kavala | 1987 | Palaz | |||||
| BPIC 1436c | Kavala | 1990 | Palaz | |||||
| BPIC 703cd | Katerini | 1977 | Palaz | |||||
| BPIC 707c | Katerini | 1977 | Palaz | |||||
| BPIC 640cd | Kilkis | 1976 | Palaz | |||||
| BPIC 647 | Kilkis | 1976 | Palaz | |||||
| BPIC 649 | Kilkis | 1976 | Palaz | |||||
| BPIC 659 | Kilkis | 1976 | Palaz | |||||
| BPIC 665 | Kilkis | 1976 | Palaz | |||||
| BPIC 667 | Kilkis | 1976 | Palaz | |||||
| BPIC F1 3 | Kilkis | 1976 | Palaz | |||||
| BPIC 1077cd | Kilkis | 1987 | Tonda Rossa | |||||
| BPIC 1078c | Kilkis | 1986 | Tonda Rossa | |||||
| Strains from Italy | ||||||||
| ISPaVe 012cd | Latium-Rome | 1991 | Tonda Gentile Romana | |||||
| ISPaVe 013cd | Latium-Rome | 1992 | Tonda Gentile Romana | |||||
| ISPaVe 037c | Latium-Rome | 1992 | Tonda Gentile Romana | |||||
| ISPaVe 038cd | Latium-Rome | 1993 | Tonda Gentile Romana | |||||
| ISPaVe 369cd | Latium-Rome | 1995 | Tonda Gentile Romana | |||||
| ISPaVe 436 | Latium-Rome | 1995 | Tonda Gentile Romana | |||||
| ISPaVe 439 | Latium-Rome | 1995 | Tonda Gentile Romana | |||||
| ISF Lab 1cd | Latium-Rome | 1997 | Tonda Gentile Romana | |||||
| ISF Lab 2c | Latium-Rome | 1997 | Tonda Gentile Romana | |||||
| ISF Lab 3c | Latium-Rome | 1997 | Tonda Gentile Romana | |||||
| ISF Lab 4 | Latium-Rome | 1997 | Tonda Gentile Romana | |||||
| ISF Lab 5 | Latium-Rome | 1997 | Tonda Gentile Romana | |||||
| ISF Lab 6 | Latium-Rome | 1998 | Tonda Gentile Romana | |||||
| ISF Lab 7 | Latium-Rome | 1998 | Tonda Gentile Romana | |||||
| ISF Lab 8 | Latium-Rome | 1998 | Tonda Gentile Romana | |||||
| ISF V 1cd | Latium-Rome | 1999 | Tonda Gentile Romana | |||||
| ISF V 2c | Latium-Rome | 1999 | Tonda Gentile Romana | |||||
| ISF V 3c | Latium-Rome | 1999 | Tonda Gentile Romana | |||||
| ISF Morcd | Latium-Rome | 1998 | Mortarella | |||||
| ISPaVe 041cd | Latium-Viterbo | 1992 | Tonda Gentile Romana | |||||
| ISPaVe 042c | Latium-Viterbo | 1992 | Tonda Gentile Romana | |||||
| ISPaVe 038cd | Latium-Viterbo | 1993 | Tonda Gentile Romana | |||||
| ISPaVe 039c | Latium-Viterbo | 1993 | Tonda Gentile Romana | |||||
| ISPaVe 040 | Latium-Viterbo | 1993 | Tonda Gentile Romana | |||||
| ISPaVe 2056c | Latium-Viterbo | 1994 | Tonda Gentile Romana | |||||
| ISPaVe 2057 | Latium-Viterbo | 1994 | Tonda Gentile Romana | |||||
| ISPaVe 2058 | Latium-Viterbo | 1994 | Tonda Gentile Romana | |||||
| ISPaVe 2059cd | Latium-Viterbo | 1994 | Tonda Gentile Romana | |||||
| ISPaVe 683c | Latium-Viterbo | 1996 | Tonda Gentile Romana | |||||
| ISPaVe 689cd | Latium-Viterbo | 1996 | Tonda Gentile Romana | |||||
| ISPaVe 690 | Latium-Viterbo | 1996 | Tonda Gentile Romana | |||||
| ISPaVe 691 | Latium-Viterbo | 1996 | Tonda Gentile Romana | |||||
| ISF Rad 1cd | Latium-Viterbo | 1998 | Tonda Gentile Romana | |||||
| ISF Rad 2 | Latium-Viterbo | 1998 | Tonda Gentile Romana | |||||
| ISF Rad 3 | Latium-Viterbo | 1998 | Tonda Gentile Romana | |||||
| ISF Rad 4 | Latium-Viterbo | 1998 | Tonda Gentile Romana | |||||
| ISF Bar 1cd | Latium-Viterbo | 1998 | Barrettona | |||||
| ISF Bar 2 | Latium-Viterbo | 1998 | Barrettona | |||||
| ISF Bar 3 | Latium-Viterbo | 1998 | Barrettona | |||||
| ISF Bar 4 | Latium-Viterbo | 1998 | Barrettona | |||||
| ISF Noccd | Latium-Viterbo | 1998 | Nocchione | |||||
| ISF VM 1cd | Latium-Viterbo | 1999 | Tonda di Giffoni | |||||
| ISF VM 2 | Latium-Viterbo | 1999 | Tonda di Giffoni | |||||
| ISF VM 3 | Latium-Viterbo | 1999 | Tonda di Giffoni | |||||
| ISF CId | Latium-Viterbo | 1999 | Tonda Gentile Romana | |||||
| ISPaVe 592cd | Piedmont | 1995 | Tonda Gentile Langhe | |||||
| ISPaVe 593 | Piedmont | 1995 | Tonda Gentile Langhe | |||||
| ISPaVe 595 | Piedmont | 1995 | Tonda Gentile Langhe | |||||
| ISPaVe 596 | Piedmont | 1995 | Tonda Gentile Langhe | |||||
| ISPaVe 598 | Piedmont | 1995 | Tonda Gentile Langhe | |||||
| ISPaVe 599cd | Piedmont | 1995 | Tonda Gentile Langhe | |||||
| ISF Lan 1cd | Piedmont | 1997 | Tonda Gentile Langhe | |||||
| ISF Lan 2 | Piedmont | 1997 | Tonda Gentile Langhe | |||||
| ISF Lan 4 | Piedmont | 1997 | Tonda Gentile Langhe | |||||
| ISF Lan 5 | Piedmont | 1997 | Tonda Gentile Langhe | |||||
| ISF Cn 1c,d | Piedmont | 1998 | Tonda Gentile Langhe | |||||
| ISF Cn 2 | Piedmont | 1998 | Tonda Gentile Langhe | |||||
| ISF Cn 3 | Piedmont | 1998 | Tonda Gentile Langhe | |||||
| ISF Cn 4 | Piedmont | 1998 | Tonda Gentile Langhe | |||||
| ISF To 1cd | Piedmont | 1998 | Tonda Gentile Langhe | |||||
| ISF To 2 | Piedmont | 1998 | Tonda Gentile Langhe | |||||
| ISF To 3 | Piedmont | 1998 | Tonda Gentile Langhe | |||||
| ISF To 4 | Piedmont | 1998 | Tonda Gentile Langhe | |||||
| ISF Cr 1cd | Piedmont | 1998 | Tonda Gentile Langhe | |||||
| ISF Cr 2 | Piedmont | 1998 | Tonda Gentile Langhe | |||||
| ISF Cr 3 | Piedmont | 1998 | Tonda Gentile Langhe | |||||
| ISF Cr 4 | Piedmont | 1998 | Tonda Gentile Langhe | |||||
| ISF P 1cd | Piedmont | 1999 | Tonda Gentile Langhe | |||||
| ISF P 2 | Piedmont | 1999 | Tonda Gentile Langhe | |||||
| ISF P 3 | Piedmont | 1999 | Tonda Gentile Langhe | |||||
| ISF P 4 | Piedmont | 1999 | Tonda Gentile Langhe | |||||
| ISF C 2cd | Campania | 1997 | Camponica | |||||
| ISF C 3cd | Campania | 1997 | Camponica | |||||
| ISF C 4cd | Campania | 1997 | Camponica | |||||
| ISF C 5 | Campania | 1997 | Camponica | |||||
| ISF C 6 | Campania | 1997 | Camponica | |||||
| ISF C 7 | Campania | 1997 | Camponica | |||||
| ISF C 8 | Campania | 1997 | Camponica | |||||
| ISF Sar A 1c,d | Sardinia | 1998 | Nocciola Sarda Piccola | |||||
| ISF Sar A 2cd | Sardinia | 1998 | Nocciola Sarda Piccola | |||||
| ISF Sar A 3 | Sardinia | 1998 | Nocciola Sarda Piccola | |||||
| ISF Sar A 4 | Sardinia | 1998 | Nocciola Sarda Piccola | |||||
| ISF Sar A 5 | Sardinia | 1998 | Nocciola Sarda Piccola | |||||
| ISF Sar A 6 | Sardinia | 1998 | Nocciola Sarda Piccola | |||||
| ISF Sar B 1cd | Sardinia | 1998 | Nocciola Sarda | |||||
| ISF Sar B 2cd | Sardinia | 1998 | Nocciola Sarda | |||||
| ISF Sar B 3 | Sardinia | 1998 | Nocciola Sarda | |||||
| ISF Sar B 4 | Sardinia | 1998 | Nocciola Sarda | |||||
| ISF Sar B 5 | Sardinia | 1998 | Nocciola Sarda | |||||
| ISF Sar B 6 | Sardinia | 1998 | Nocciola Sarda | |||||
| ISF Sar C 1cd | Sardinia | 1998 | Tonda Gentile Langhe | |||||
| ISF Sar C 2cd | Sardinia | 1998 | Tonda Gentile Langhe | |||||
| ISF Sar C 3 | Sardinia | 1998 | Tonda Gentile Langhe | |||||
| ISF Sar C 4 | Sardinia | 1998 | Tonda Gentile Langhe | |||||
| ISF Sar C 5 | Sardinia | 1998 | Tonda Gentile Langhe | |||||
| ISF Sar C 6 | Sardinia | 1998 | Tonda Gentile Langhe | |||||
| ISF Sic 1cd | Sicily | 1999 | Santa Maria del Gesù | |||||
| ISF Sic 2cd | Sicily | 1999 | Santa Maria del Gesù | |||||
| ISF Sic 3cd | Sicily | 1999 | Santa Maria del Gesù | |||||
| ISF Sic 4 | Sicily | 1999 | Santa Maria del Gesù | |||||
| ISF Sic 5 | Sicily | 1999 | Santa Maria del Gesù | |||||
| P. syringae pv. syringae strains | ||||||||
| NCPPB 281Tc | United Kingdom | 1950 | Syringa vulgaris | |||||
| NCPPB 1092c | New Zealand | 1951 | Prunus armeniaca | |||||
| NCPPB 1093c | New Zealand | 1951 | Prunus armeniaca | |||||
| NCPPB 1087c | Hungary | 1961 | Prunus avium | |||||
| NCPPB 1652c | South Africa | 1957 | Pisum sativum | |||||
| NCPPB 2434c | United States | 1971 | Nerium oleander | |||||
| PD 2618c | United States | NKe | Hordeum vulgare | |||||
| P. syringae pv. eriobotryae NCPPB 2331Tc | United States | 1970 | Eriobotrya japonica | |||||
| P. syringae pv. helianthi NCPPB 2640Tc | Mexico | 1972 | Helianthus annuus | |||||
| P. syringae pv. lachrymans PD 904c | The Netherlands | 1987 | Cucumis sativus | |||||
| P. syringae pv. morsprunorum NCPPB 2787c | Greece | 1971 | Prunus avium | |||||
| P. syringae pv. persicae NCPPB 2761Tc | France | 1974 | Prunus persica | |||||
| P. syringae pv. ulmi NCPPB 632Tc | Yugoslavia | 1958 | Ulmus sp. |
NCPPB, National Collection of Plant Pathogenic Bacteria, York, United Kingdom; BPIC, Culture Collection of Benaki Phytopathological Institute, Kiphissia-Athens, Greece; ISPaVe, Culture Collection of Istituto Sperimentale per la Patologia Vegetale, Rome, Italy; ISF, Culture Collection of Istituto Sperimentale per la Frutticoltura, Rome, Italy; PD, Culture Collection of Plant Protection Service, Wageningen, The Netherlands.
T = type strain.
Strain with which ARDRA was performed.
Strain with which pathogenicity tests were performed.
NK, not known.
Survey and isolation of field strains.
Field surveys were carried out in the main hazelnut-growing areas of Italy, including Campania, Latium, Piedmont, Sardinia, and Sicily. Samples of diseased twigs or branches were collected at sites where partial or total wilting of trees was noticed. The samples were immediately cooled at 4 to 5°C, and they were processed for isolation within 48 h after removal. The surveys allowed us to ascertain the current status of hazelnut decline in these regions. The techniques used previously were used for isolation (15, 16, 19). The epidermis of a twig or branch was aseptically removed, and small portions of the twig or branch (length, 3 to 5 mm) were macerated in sterile physiological saline (SPS) (0.85% NaCl in distilled water). Then the suspensions were serially diluted, and 0.1-ml aliquots were spread onto NSA. The plates were incubated at 25 to 27°C for 3 to 4 days. The strategy used to characterize the strains included the following steps: (i) collecting isolates from symptomatic hazelnut orchards in different geographical areas; (ii) grouping these isolates and previously isolated strains on the basis of an analysis of the restriction patterns of 16S DNAs amplified by PCR (ARDRA); (iii) differentiating the populations at the strain level by repetitive PCR fingerprinting, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of whole-cell proteins, analysis of the utilization of carbohydrates and related carbon compounds, and analysis for the presence of the syrB gene; (iv) clustering the isolates by the unweighted pair group method using aritmethic averages (UPGMA); and (v) performing, with representative strains of the groups, pathogenicity tests with hazelnut and other plant species in order to assess specificity and virulence.
ARDRA.
The basic ARDRA technique described by Vaneechoutte et al. (22) was utilized. The DNAs coding for the 16S rRNAs of strains were amplified with primers PO (5′-GAGAGTTTGATCCTGGCTCAG) and P6 (5′-CTACGGCTACCTTGTTACGA). These primers were designed by Grifoni et al. (4) on the basis of the conserved bacterial sequences at the 5′ and 3′ ends of the 16S rRNA gene (positions 27f and 1495r in Escherichia coli rDNA, respectively) and allowed amplification of almost the entire gene. To prepare total genomic DNA, a modification of the techniques used by Smith et al. (20) was used. A loopful (diameter, approximately 3 mm) of a single colony of each strain that had been grown for 24 h on NSA at 25 to 27°C was suspended in SPS and centrifuged at 12,000 × g for 2 min. After the supernatant was discarded, the pellet was suspended in bidistilled, filtered, sterilized water to an optical density corresponding to 1 × 108 to 2 × 108 CFU ml−1. The suspension was placed in boiling water for 10 min and then stored at −20°C. Each 50-μl (final volume) PCR mixture contained 6 μl of lysed cell suspension, Promega Taq buffer (1.5 mM MgCl2), each deoxynucleoside triphosphate at a concentration of 200 μM, 36 pmol of each primer, and 1.5 U of Taq DNA polymerase (Promega). The mixture was overlaid with 50 μl of mineral oil. The reaction mixtures were incubated on an MJ Research PTC 100 programmable thermal controller at 95°C for 2 min and then subjected to 35 cycles consisting of 95°C for 30 s, the annealing temperature for 30s, and 72°C for 4 min. The annealing temperature was 60°C for the first 5 cycles, 55°C for the next 5 cycles, and 50°C for the last 25 cycles. Finally, the mixtures were incubated at 72°C for 10 min and then at 60°C for 10 min. Two microliters of each amplification mixture was analyzed by agarose (1.0%, wt/vol) gel electrophoresis in 0.5× Tris-borate-EDTA (TBE) buffer (80 mM Tris borate, 89 mM boric acid, 2 mM EDTA; pH 8) containing 0.5 μg of ethidium bromide per ml at 5.0 V cm−1. A 5-μl aliquot of each PCR mixture containing approximately 1.5 μg of amplified 16S rDNA was digested with the following restriction endonucleases as recommended by the manufacturer (Boehringer Mannheim): CfoI, DraI, EcoRI, HaeIII, HinfI, MvaI, PstI, RsaI, and TaqI. The reaction products were analyzed by agarose (2.5%, wt/vol) gel electrophoresis in 0.5× TBE buffer containing 0.5 μg of ethidium bromide per ml. Restriction fragment profiles were used to determine genetic relationships among the strains. Pairwise comparisons were made between all strains to generate a similarity matrix based on the presence (1) or absence (0) of a specific band. Cluster analysis with UPGMA was used to identify genetically similar groups by using the NTSYS software, version 1.80 (Exeter Software, New York, N.Y.). Type and reference strains of Pseudomonas syringae (Table 1) were also included in the comparison as outgroups.
Repetitive PCR genomic fingerprinting.
The repetitive PCR method used was that of Louws et al. (9). The ERIC, BOX, and REP primer sets were synthesized by Eurogentech. Amplification was performed on an MJ Research PTC 100 programmable thermal controller. Each 25-μl reaction mixture contained each deoxynucleoside triphosphate at a concentration of 200 μM, 2 mM MgCl2, 60 pmol of each primer, 1.0 U of Taq polymerase, and 3 μl of a cell DNA preparation. The PCR mixture was overlaid with 25 μl of mineral oil. The thermal cycling procedure was that used by Louws et al. (9). Subsequently, the PCR amplification products were separated by gel electrophoresis on a 2.0% agarose gel in 1× Tris-acetic acid-EDTA buffer at 5 V cm−1 for 5 h, stained with ethidium bromide, visualized with a UV transilluminator, and photographed with Polaroid type 55 film. The PCR amplifications were performed in duplicate. The method of Smith et al. (20) was used for gel analysis. The gels were examined, and bands present on both amplification gels were scored and recorded. Similarity coefficients for all pairwise combinations were determined by using Dice's coefficients (3) and were clustered by UPGMA by using the NTSYS software (version 1.80). Repetitive PCR was also performed with the P. syringae strains used as outgroups for ARDRA.
Whole-cell protein analysis.
Soluble whole-cell protein extracts were obtained for each strain as described by Vauterin et al. (23). SDS-PAGE was performed by using a Bio-Rad Mini Protean apparatus and 12% (wt/vol) polyacrylamide gels that were electrophoresed vertically at 40 mA (constant current) and 4°C until the bromophenol blue tracking dye had migrated to the bottom. The gels were stained for 1 h at 24°C with a staining solution containing 500 ml of methanol, 100 ml of acetic acid, 125 ml of stain stock (10 g of Coomassie blue R250 in 500 ml of bidistilled water), and 275 ml of bidistilled water. Destained gels were photographed, and the gels were examined. Each strain was examined in duplicate. The method described above for PCR genomic fingerprinting was used for gel analysis.
Detection of syrB gene, biochemical tests, and carbon compound utilization analysis.
The presence of the syrB gene was determined by using the procedures of Sorensen et al. (21). Primers B1 and B2 were synthesized by Eurogentech (Seraing, Belgium).
Before the abilities of all strains to utilize different carbon compounds were assessed, we performed the following biochemical tests to distinguish fluorescent plant-pathogenic pseudomonads by using the methods described by Lelliott and Stead (8): tests for levan production, oxidase activity, potato soft rot, arginine dihydrolase activity, and tobacco hypersensitivity (LOPAT tests). Production of fluorescent pigments was determined by using medium B of King et al. (6). The type of metabolism (oxidative or fermentative) was also assessed. In addition, the following tests were performed: tests for gelatin liquefaction, esculin and arbutin hydrolysis, production of tyrosinase, and utilization of inositol, sorbitol, quinate, anthranilate, l-(−)-homoserine, and l-(+)-tartaric acid. Possible utilization of the following carbon compounds was also tested: l-(−)-sorbose, fructose, l-(+)-ramnose, cellobiose, ribose, d-(+)-melibiose, malic acid, histidine, adenine, guanine, l-alanine, tymine, and trigonelline. The basal medium of Ayers et al. (2) was used, and growth on the surface of the agar was considered an indication that a substrate was utilized.
Virulence and host specificity assessment.
After strains representing each group, each site, and each year of isolation were clustered, pathogenicity tests were performed in order to assess the specificity and virulence of each strain (Table 1). Hazelnut trees were each inoculated in early autumn through leaf scars located midway between the ends of a 1-year-old twig that was 30 to 40 cm long. This method has repeatedly been shown to be very effective for artificial inoculation and disease development (15, 16, 19). Adult C. avellana cv. Tonda Gentile Romana and C. avellana cv. Tonda Gentile delle Langhe trees were used as susceptible hosts. For each strain, 10-fold dilutions from 107 to 1 × 103 CFU ml −1 were prepared. For each strain, 10 μl of each suspension was placed with a micropipette on the surface of a leaf scar immediately after removal of the leaf at the base of the petiole. Ten leaf scars per strain per suspension, randomly distributed in the crown, were inoculated at the beginning of October. Control plants were inoculated in the same way with only SPS. The virulence of each strain was assessed the following May by recording (i) the number of completely wilted twigs and (ii) the number of dead plants. After transformation of the percentages to angular values, the data were statistically analyzed by using the Student t test to compare two different populations at a time. Reisolation from symptomatic plants was performed in May by using the techniques described above. To test the host specificities of the microorganisms, other plant species, including Prunus armeniaca, Prunus persica, Prunus salicina, and Pyrus communis, were inoculated either in the early autumn or in the spring by using the same strains, the same inoculum concentrations, and the same technique. In addition, in order to test plant species commonly infected by the ubiquitous organism P. syringae pv. syringae, representative strains were also inoculated at concentrations of 1 × 106 to 2 × 106 CFU ml−1 into orange and lemon fruits, pear fruitlets, and bean pods, as well as along petioles and into leaves of Syringa vulgaris (25) and Zea mays.
RESULTS
Isolation of pseudomonads from hazelnut twigs.
NSA allowed us to recover levan-positive colonies that showed weak fluorescence after restreaking on medium B of King et al. All isolates had respiratory metabolism and showed the following responses in the LOPAT tests: levan positive, oxidase negative, potato soft rot negative, arginine dehydrolase negative, and tobacco hypersensitivity positive (group Ia).
ARDRA.
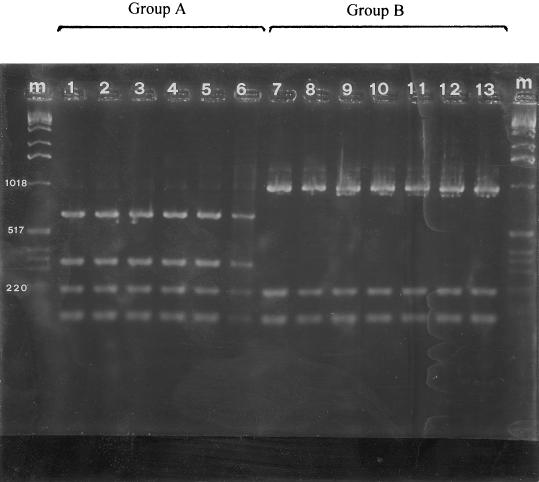
A total of 53 strains representing each geographical area, each field site, and each time of isolation were analyzed by ARDRA. The rDNA-based primers allowed amplification of 16S rDNA from all 53 strains tested. The PCR products included a single band at about 1,500 bp. All of the amplified 16S rDNAs were digested with the nine restriction endonucleases used. Depending on the restriction endonuclease, one to five restriction fragments were observed. However, it was possible to discriminate among the strains isolated at different times from different geographical areas and sites only with HinfI (Fig 1). UPGMA cluster analysis revealed that two groups of ARDRA patterns were obtained with HinfI that were related at a genetic similarity level of 57%. Group A included all of the strains isolated from different sites in northern Greece and from different sites in central Italy (i.e., the province of Viterbo and one orchard in the province of Rome established with propagative material obtained from Viterbo). Group B included all of the strains isolated from the other regions of Italy (i.e., Campania, Latium, Piedmont, Sardinia, and Sicily), and it was not possible to further differentiate among sites and time of isolation. Also, the P. syringae strains produced patterns identical to those produced by group A and B strains when they were tested with eight of the nine restriction endonucleases. With HinfI they produced the same restriction patterns as the group B strains.
FIG. 1.
ARDRA band patterns for amplified 16S rDNA obtained by using HinfI restriction endonuclease for P. avellanae (lanes 1 to 6) and fluorescent pseudomonads pathogenic for C. avellana (lanes 7 to 13). Lanes m, molecular size marker (1-kb ladder; Gibco-BRL). lane 1, ISPaVe 012; lane 2, ISPaVe 056; lane 3, ISPaVe 2059; lane 4, BPIC 631; lane 5, BPIC 703; lane 6, BPIC 1077; lane 7, ISPaVe 592; lane 8, ISF Lan 8; lane 9, ISF C4; lane 10, ISF Lab 6; lane 11, ISF Sic 3; lane 12, ISF Sar C5; lane 13, ISF Sar A1.
Repetitive PCR analysis.
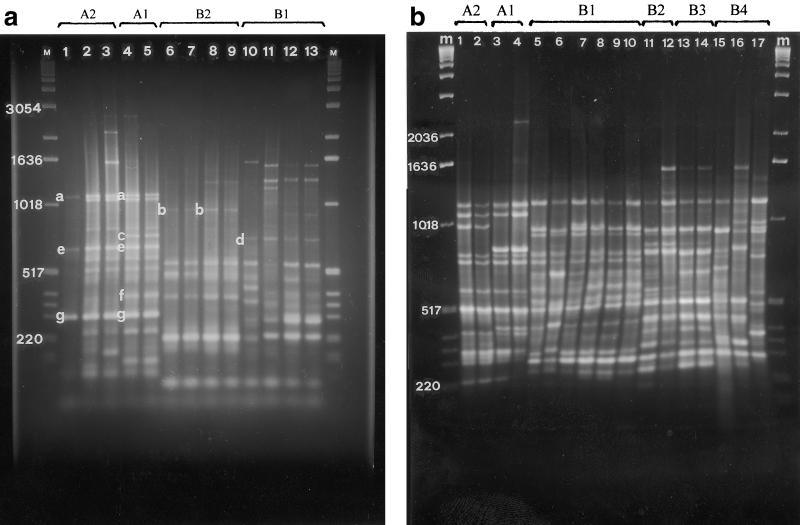
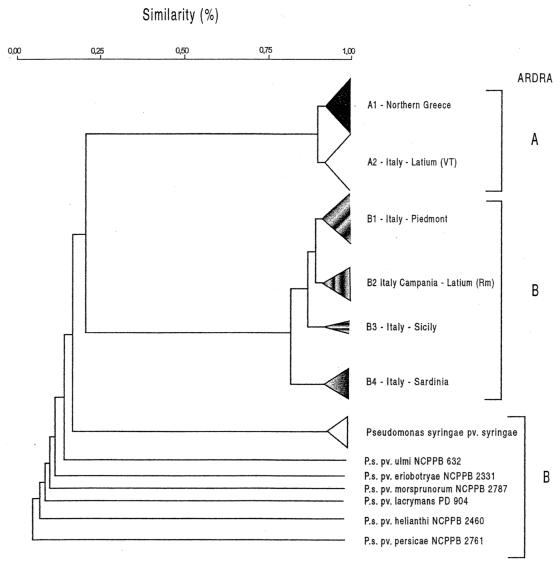
All 118 strains shown in Table 1 were assessed with ERIC, BOX, and REP primers in repetitive PCR experiments. All primer sets gave reproducible genomic PCR profiles consisting of bands ranging in size from approximately 100 bp to 3 kb. For the UPGMA analysis a total of 43 reproducible clearly resolved bands were scored; 19 bands were scored for the ERIC primer set, 18 bands were scored for the BOX primer set, and 6 bands were scored for the REP primer set. The ERIC and BOX primer sets were more discriminative than the REP primer set for differentiating the strains. With the ERIC primers distinct DNA polymorphism was observed in the region between 100 and 1,600 bp (Fig. 2a), whereas with the BOX primers polymorphism was evident in the region between 200 and 2,300 bp (Fig. 2b); also, the REP primers were more discriminative in the latter region. UPGMA cluster analysis of the combined data obtained in the ERIC, BOX, and REP PCR experiments revealed that the pathogenic fluorescent pseudomonads associated with hazelnut could be separated into two main groups that were related at a level of similarity of approximately 20% (Fig. 3). Group A included all of the strains isolated in northern Greece and the strains from central Italy (i.e., the province of Viterbo and the strains from one orchard in the province of Rome established with propagative material obtained from the province of Viterbo). This group could be further differentiated into the following two subgroups: subgroup A1, which included the strains from northern Greece; and subgroup A2, which included the strains from central Italy (Fig. 2a). These subgroups were related at a similarity level of 90%. The members of subgroup A1 produced two differentiating bands at around 300 and 800 bp (Fig. 2a). Group B included the strains isolated in Piedmont, Campania, Sardinia, and Sicily and from hazelnut orchards in Latium in the province of Rome. For this group, cluster analysis indicated that the strains could be differentiated on the basis of geographic area into four different subgroups, subgroups B1, B2, B3, and B4 (Table 2). In fact, strains from Piedmont, Campania, Latium, Sicily, and Sardinia can be clearly differentiated from each other on the basis of representative bands (Fig. 2). Within the subgroups, the strains from Campania and Latium (subgroup B2) and the strains from Sicily (subgroup B3) are homogeneous, whereas the strains from Piedmont (subgroup B1) and the strains from Sardinia (subgroup B4) are heterogeneous. Interestingly, strains from Campania and strains from Latium, which were isolated from different hazelnut cultivars and from sites that were more than 200 km apart, produced the same genomic pattern. The strains of the P. syringae pathovars produced different profiles than the strains isolated from hazelnut produced.
FIG. 2.
(a) Repetitive PCR fingerprint patterns for genomic DNAs of pseudomonad strains pathogenic for C. avellana obtained by using the ERIC primer set. Lanes M, molecular size marker (1-kb ladder; Gibco-BRL); lane 1, ISPaVe 036; lane 2, ISPaVe 2059; lane 3, ISPaVe 691; lane 4, BPIC 631; lane 5, BPIC 1078; lane 6, ISF C 2; lane 7, ISF C 3; lane 8, ISF Lab 1; lane 9, ISF Lab 2; lane 10, ISPaVe 595; lane 11, ISPaVe 592; lane 12, ISF Lan 6; lane 13, ISF Lan 8. The letters indicate the bands that differentiate the subgroups, as follows: subgroup A2, bands a, e, and g; subgroup A1, bands a, c, e, f, and g; subgroup B2, band b; and subgroup B1, band d. (b) Repetitive PCR fingerprint patterns obtained for genomic DNAs of pseudomonad strains pathogenic for C. avellana obtained by using the BOX primer set. Lanes m, molecular size marker (1-kb ladder; Gibco-BRL); lanes 1 and 2, subgroup A2 (lane 1, ISPaVe 012; lane 2, ISPaVe 056); lanes 3 and 4, subgroup A1 (lane 3, BPIC 703; lane 4, BPIC 1077); lanes 5 to 10, subgroup B1 (lane 5, ISPaVe 592; lane 6, ISPaVe 595; lane 7, ISF Lan 8; lane 8, ISF Cr 1; lane 9, ISF Cr 6; lane 10, ISF To 3); lanes 11 and 12, subgroup B2 (lane 11, ISF C 4; lane 12, ISF Lab 6); lanes 13 and 14, subgroup B3 (lane 13, ISF Sic 3; lane 14, ISF Sic 4); lanes 15 to 17, subgroup B4 (lane 15, ISF Sar C6; lane 16, ISF Sar B5; lane 17, ISF Sar A1).
FIG. 3.
Simplified dendrogram showing relationships among 118 fluorescent pseudomonad strains pathogenic for C. avellana constructed by using the results of repetitive PCR performed with ERIC, BOX, and REP primer sets. Cluster analysis was performed by UPGMA with a matrix calculated with Dice's coefficients. VT, province of Viterbo in Latium; Rm, Rome; P.s., P. syringae. Subgroups A1 and A2 contained strains belonging to P. avellanae.
TABLE 2.
Main genetic, phenotypic, and pathological features of fluorescent pseudomonads pathogenic for C. avellana isolated in northern Greece and different geographic areas of Italy
| Country (region) | Group or subgroupa
|
syrB | |||
|---|---|---|---|---|---|
| ARDRA | Repetitive PCR | Protein | Utilization of compounds and virulenceb | ||
| Northern Greece | A | A1 | A1 | A | − |
| Italy (Viterbo) | A | A2 | A2 | A | − |
| Italy (Piedmont) | B | B1 | B1 | B | − |
| Italy (Latium) | B | B2 | B2 | B | + |
| Italy (Campania) | B | B2 | B2 | B | + |
| Italy (Sicily) | B | B3 | B2 | B | + |
| Italy (Sardinia) | B | B4 | B3 | B | + |
Group A includes P. avellanae strains.
The group B strains utilized arbutin, esculin, gelatin, l-histidine, dl-homoserine, quinic acid, sorbitol, trigonelline, and tyrosinase. The group A strains did not utilize these compounds.
Whole-cell protein analysis.
Duplicate protein electrophoretograms were obtained for the 118 strains listed in Table 1. The reproducibility was excellent, and the profiles were judged to be good enough for cluster analysis. Additionally, representative bands (12 clearly resolved bands between 21.5 and 66.2 kDa) were selected for UPGMA analysis. Consistent with the repetitive PCR results, this UPGMA analysis revealed two major groups of strains related at a similarity level of 65%. Group A included strains from northern Greece and central Italy; whereas group B included all of the other strains isolated in Campania, Latium, Piedmont, Sardinia, and Sicily. Also, two subgroups were differentiated in group A; subgroup A1 contained strains from northern Greece, and subgroup A2 contained the strains obtained from central Italy (i.e., the province of Viterbo and the orchard in the province of Rome established with propagative material from Viterbo). Two bands at around 40 and 60 kDa differentiated the two subgroups. Within the subgroups, no differences in the protein profiles of the strains were observed. Group B included the following three subgroups: subgroup B1, which contained strains isolated in Piedmont; subgroup B2, which contained strains isolated in Campania, Latium, Sicily; and subgroup B3, which contained strains isolated in Sardinia. Heterogeneity was observed only in the case of the strains obtained from Piedmont and Sardinia. The major differences in group A were differences at molecular masses between 31 and 66.2 kDa.
Detection of syrB gene, biochemical tests, and carbon compound utilization.
PCR amplification with primers B1 and B2 was performed with all 118 strains tested, and only the strains isolated in Campania and Latium (i.e., the province of Rome) (subgroup B2), Sicily (subgroup B3), and Sardinia (subgroup B4) produced the 752-bp band which indicated that the syrB gene was present (Table 2). All 118 strains tested produced the same results in the LOPAT tests (i.e., group Ia). All strains produced the following results for utilization of carbon compounds: they were negative for utilization of l-(−)-sorbose, l-(+)-rhamnose, cellobiose, d-(+)-melibiose, l-(+)-tartaric acid, adenine, guanine, thymine, and anthranilic acid, whereas they were positive for utilization of d-(−)-fructose, ribose, meso-inositol, and l-alanine. The group A strains were negative for the following tests: arbutin and esculin hydrolysis, gelatin liquefaction, production of tyrosinase, and utilization of sorbitol, quinic acid, dl-homoserine, trigonelline, and l-histidine. All group B strains were positive for these tests (Table 2).
Virulence and host specificity.
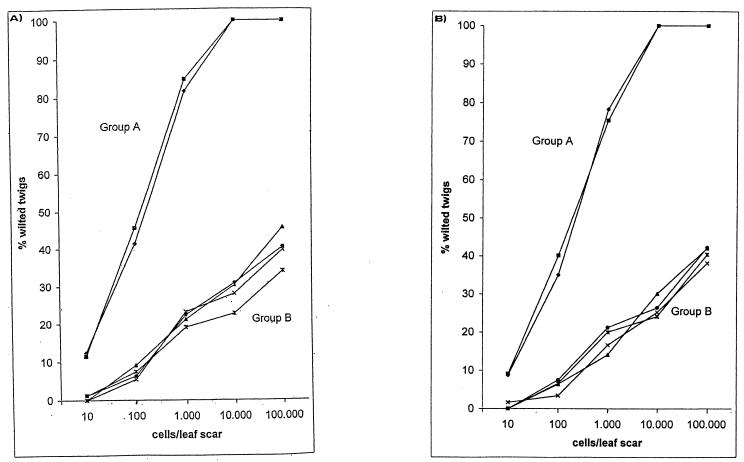
Inoculation of 1-year-old twigs in the early autumn through leaf scars proved to be a satisfactory test for determining the differences in virulence of the pseudomonads pathogenic for hazelnut. Within the geographic area where hazelnut is cultivated, all of the strains used for the test produced similar responses. They caused wilting of the twigs that varied according to the dose and the virulence of the organism, and it was possible to differentiate two major groups of strains on this basis. All of the strains isolated in northern Greece and central Italy (i.e., the province of Viterbo and the orchard in the province of Rome established with propagative material obtained from Viterbo) (group A) were much more virulent than the other strains and produced complete wilting of many twigs after 7 months. Indeed, with cultivars Tonda Gentile Romana and Tonda Gentile delle Langhe such strains caused wilting of 100% of the twigs when a dose of around 104 or 105 cells per leaf scar was used (Fig. 4). In addition, approximately 50% of the plants inoculated through leaf scars with such doses died during the summer. In contrast, inoculation of all of the strains isolated in the other regions (group B) at the highest doses used resulted in complete wilting of no more than 45.6% of the twigs, and none of the inoculated plants died during the summer. When the inoculum was around 10 cells per leaf scar, only the strains from northern Greece and central Italy caused wilting of some twigs (Fig. 4). The results obtained for the two groups were significantly different (P < 0.01, as determined by Student's t test). The assessment of virulence also revealed that strains isolated in the same area (i.e., the province of Viterbo) from different hazelnut cultivars exhibited similar virulence characteristics when they were inoculated into the cultivar from which they were isolated and when they were inoculated into another cultivar. In addition, the virulence results confirmed that strains isolated from cultivar Tonda Gentile Romana plants growing in two different areas of Latium were different. No symptoms were observed in the control-treated twigs. Reisolation of both cultivars from inoculated plants yielded bacterial colonies on NSA that were confirmed to be group A or group B colonies by biochemical tests and repetitive PCR fingerprinting. When inoculated into other plant species, group A and subgroup B1 strains did not produce any symptoms, whereas subgroup B2, B3, and B4 strains caused necrotic lesions around the inoculation sits on orange fruits, bean pods, and Z. mays leaves. Based on the results obtained by ARDRA, repetitive PCR genomic fingerprinting and UPGMA analysis, whole-cell protein analysis, carbon compound utilization and host specificity analyses, and syrB gene analysis, we concluded that the group B strains are similar to P. syringae pv. syringae.
FIG. 4.
Virulence expressed as percentages of wilted twigs after inoculation of P. avellanae strains (group A) and other fluorescent pseudomonads (group B) pathogenic for C. avellana cv. Tonda Gentile Romana (A) and C. avellana cv. Tonda Gentile delle Langhe (B). Bacterial suspensions in sterile saline were photometrically adjusted to an optical density corresponding to 1 × 107 CFU ml−1. Serial 10-fold dilutions up to 1 × 103 CFU ml−1 were prepared. Ten microliters of a suspension was placed on each leaf scar. Symbols: ▪, subgroup A1; ⧫, subgroup A2; ×, subgroup B1; ▴, subgroup B2; •, subgroup B3; X, subgroup B4.
DISCUSSION
In this study we ascertained that two groups of fluorescent pseudomonads are currently causing disease in C. avellana in Italy. The results obtained with genetic and phenotypic techniques in this study are in agreement with the results of an assessment of the virulence of the strains and clearly differentiate group A from group B. In fact, when ARDRA and restriction endonuclease HinfI were used, the genetic relatedness between the two groups was 57%. The other eight endonucleases used did not separate the strains. Group A included the typical P. avellanae strains isolated in northern Greece, in the province of Viterbo (central Italy), and in one hazelnut orchard in the province of Rome established with propagative material obtained from Viterbo. Repetitive PCR and whole-cell protein profiling differentiated group A into two subgroups, subgroup A1 (strains from northern Greece) and subgroup A2 (strains from central Italy). In fact, PCR products that were around 300 and 800 bp long, were obtained only with the strains from northern Greece, and were obtained with the ERIC primer set and protein bands at around 40 and 60 kDa clearly indicated that these subgroups could be differentiated. Within the subgroups, the strains seemed to be heterogeneous both in terms of space and in terms of time, even though very often the variability was revealed by only one PCR product. However, to verify the population structures of the different subgroups, an in-depth study with multilocus enzyme electrophoresis (10, 11) is under way.
Repetitive PCR and biochemical and pathogenicity tests indicated that the group B strains were similar to P. syringae pv. syringae. The carbon compound utilization tests showed that these strains could be differentiated easily from P. avellanae. In fact, all group B strains utilized quinic acid, sorbitol, dl-homoserine, l-histidine, and trigonelline. In addition, they liquefied gelatin and were esculin and tyrosinase positive. In contrast, all P. avellanae strains are negative in such tests. Moreover, group B differed from P. syringae pv. syringae as determined by the tests proposed by Young (25). Indeed, all group B strains utilized dl-homoserine and were tyrosinase positive, whereas the 48 P. syringae pv. syringae strains examined by Young did not utilize dl-homoserine and were tyrosinase negative (25). Furthermore, like P. avellanae, the group B strains did not produce any symptoms in lilac, lemon fruits, or pear fruitlets or in apricot, Japanese plum, peach, and walnut twigs. By contrast, they caused necrotic lesions in orange fruits, bean pods, and Z. mays leaves. Within group B, there was heterogeneity among the strains isolated in Piedmont and Sardinia. Even though all the strains were grouped together to simplify the data reported, repetitive PCR genomic fingerprinting and whole-cell protein profiling revealed that each of the populations isolated in different counties was quite heterogeneous. Actually, repetitive PCR revealed at least five populations of subgroup B1 strains in Piedmont (19), and all of these populations were mildly pathogenic for C. avellana cv. Tonda Gentile delle Langhe. Remarkably, the strains isolated from C. avellana cv. Tonda Gentile delle Langhe trees in Sardinia produced profiles that were different from the profiles produced by the strains isolated in Piedmont.
Subgroup B2 included the strains isolated in Latium and Campania from two locally cultivated hazelnut cultivars. All of the strains had identical genomic and phenotypic features, as well as similar virulence characteristics. The five strains isolated in Sicily were homogeneous and clustered separately as determined by PCR, whereas they produced the same profile as subgroup B2 strains when they were assessed by the protein profiling technique. These findings indicate that the pseudomonad populations pathogenic for C. avellana exhibit genomic and phenotypic features that may be related to the geographic area of hazelnut cultivation. In fact, in each region except Campania and Latium we found a different pattern; however, Campania and Latium are neighboring regions. Therefore, the variation in the patterns did not appear to be due to cultivar selection. The different patterns obtained for the strains isolated in Piedmont and Sardinia from C. avellana cv. Tonda Gentile delle Langhe support this hypothesis. The data may reflect the fact that hazelnut cultivation in Asia and Europe is still based on cultivars from ancient times that are adapted to certain areas where introduction of modern cultivars, obtained from breeding programs, is rare.
Our assessment of virulence clearly showed that the group A strains are very virulent and have the capacity to produce complete wilting of most artificially inoculated twigs and, in many cases, complete death of the plant within a few months after inoculation. Such a high level of virulence can explain the epidemics of hazelnut bacterial canker currently found both in northern Greece and in central Italy, where thousands of trees have already died. In contrast, group B strains are less virulent. In fact, when a high dose (100,000 cells per leaf scar) was used, none of the strains tested was able to produce wilting in more than 45.6% of the inoculated twigs, whereas each P. avellanae strain was capable of killing all the twigs of each cultivar. The fact that the group B strains were so much less virulent was consistent with the observation that in the hazelnut orchards of Campania, Latium (the province of Rome), Piedmont, Sardinia, and Sicily twig, branch, and tree dieback occurs frequently but complete and sudden death of trees in these orchards is rare. Within the fluorescent pseudomonads, four virulence groups were identified for P. syringae pv. syringae strains isolated from pear (24), whereas P. syringae pv. persicae strains can be clearly grouped based on the country of origin (26).
Finally, we found that in the Latium region contemporaneous populations of the two groups were isolated from the same cultivar. However, the orchard in which group A strains were found was recently established with propagative material obtained from the Viterbo province, where P. avellanae is widespread; in contrast, the orchards in which group B strains were isolated were established with autochtonous propagative material in the traditional manner. The orchards are 15 km apart, and they are not the same age. Such findings indicate that P. avellanae can spread latently through hazelnut suckers (15) and that repetitive PCR is a reliable technique for epidemiological studies (9). Moreover, the sudden P. avellanae epiphytotic observed in the new orchard confirmed the recent origin of the pathogen in central Italy, where group B pseudomonads have probably been present for a long time. It has to be stressed that after 10 years of research we have never found group B pseudomonads in the Viterbo area.
P. syringae pveriobotryaeThe NetherlandsisolationTonda Gentile Langhe ISF Lan 2Piedmont1997Tonda Gentile Langhe ISF Lan 4Piedmont1997Tonda Gentile Langhe ISF Lan 5Piedmont1997Tonda Gentile Langhe ISF Cn 1c,dPiedmont1998Tonda Gentile Langhe ISF Cn 2Piedmont1998Tonda Gentile Langhe ISF Cn 3Piedmont1998Tonda Gentile Langhe ISF Cn 4Piedmont1998Tonda Gentile Langhe ISF To 1c,dPiedmont1998Tonda Gentile Langhe ISF To 2Piedmont1998Tonda Gentile Langhe ISF To 3Piedmont1998Tonda Gentile Langhe ISF To 4Piedmont1998Tonda Gentile Langhe ISF Cr 1c,dPiedmont1998Tonda Gentile Langhe ISF Cr 2Piedmont1998Tonda Gentile Langhe ISF Cr 3Piedmont1998Tonda Gentile Langhe ISF Cr 4Piedmont1998Tonda Gentile Langhe ISF P 1c,dPiedmont1999Tonda Gentile Langhe ISF P 2Piedmont1999Tonda Gentile Langhe ISF P 3Piedmont1999Tonda Gentile Langhe ISF P 4Piedmont1999Tonda Gentile Langhe ISF C 2c,dCampania1997Camponica ISF C 3c,dCampania1997Camponica ISF C 4c,dCampania1997Camponica ISF C 5Campania1997Camponica ISF C 6Campania1997Camponica ISF C 7Campania1997Camponica ISF C 8Campania1997Camponica ISF Sar A 1c,dSardinia1998Nocciola Sarda Piccola ISF Sar A 2c,dSardinia1998Nocciola Sarda Piccola ISF Sar A 3Sardinia1998Nocciola Sarda Piccola ISF Sar A 4Sardinia1998Nocciola Sarda Piccola ISF Sar A 5Sardinia1998Nocciola Sarda Piccola ISF Sar A 6Sardinia1998Nocciola Sarda Piccola ISF Sar B 1c,dSardinia1998Nocciola Sarda ISF Sar B 2c,dSardinia1998Nocciola Sarda ISF Sar B 3Sardinia1998Nocciola Sarda ISF Sar B 4Sardinia1998Nocciola Sarda ISF Sar B 5Sardinia1998Nocciola Sarda ISF Sar B 6Sardinia1998Nocciola Sarda ISF Sar C 1c,dSardinia1998Tonda Gentile Langhe ISF Sar C 2c,dSardinia1998Tonda Gentile Langhe ISF Sar C 3Sardinia1998Tonda Gentile Langhe ISF Sar C 4Sardinia1998Tonda Gentile Langhe ISF Sar C 5Sardinia1998Tonda Gentile Langhe ISF Sar C 6Sardinia1998Tonda Gentile Langhe ISF Sic 1c,dSicily1999Santa Maria del Gesù ISF Sic 2c,dSicily1999Santa Maria del Gesù ISF Sic 3c,dSicily1999Santa Maria del Gesù ISF Sic 4Sicily1999Santa Maria del Gesù ISF Sic 5Sicily1999Santa Maria del Gesù P. syringae pv. syringae strains NCPPB 281TUnited Kingdom1950Syringa vulgaris NCPPB 1092New Zealand1951Prunus armeniaca NCPPB 1093New Zealand1951Prunus armeniaca NCPPB 1087Hungary1961Prunus avium NCPPB 1652South Africa1957Pisum sativum NCPPB 2434United States1971Nerium oleander PD 2618United StatesNKHordeum vulgareP. syringae pv. eriobotryae NCPPB 2331TcUnited States1970Eriobotrya japonicaP. syringae pv. helianthi NCPPB 2640TcMexico1972Helianthus annuusP. syringae pv. lachrymans PD 904The Netherlands1987Cucumis sativusP. syringae pv. morsprunorum NCPPB 2787Greece1971Prunus aviumP. syringae pv. persicae NCPPB 2761TcFrance1974Prunus persicaP. syringae pv. ulmi NCPPB 632TcYugoslavia1958Ulmus sp.
Acknowledgments
This work was financed in part by research grant 201.06.35/21.06.15 (Le batteriosi del noce e del nocciolo) from Consiglio Nazionale delle Ricerche-Comitato Scienze Agrarie.
REFERENCES
- 1.Achouak, W., L. Sutra, T. Heulin, J.-M. Meyer, N. Framin, S. Degrave, R. Christen, and L. Gardan. 2000. Pseudomonas brassicacearum sp. nov. and Psedomonas thivervalensis sp. nov., two root-associated bacteria from Brassica napus and Arabidopsis thaliana. Int. J. Syst. Bacteriol. E vol. Microbiol. 50:9-18. [DOI] [PubMed] [Google Scholar]
- 2.Ayers, S. H., P. Rupp, and W. T. Johnson. 1919. A study of the alkaly-forming bacteria in milk. U.S. Department of Agriculture Bulletin 782. Department of Agriculture, Washington, D.C.
- 3.Dice, L.R. 1945. Measurement of the amount of ecological association between species. Ecology 26:297-302. [Google Scholar]
- 4.Grifoni, A., M. Bazzicalupo, C. Di Serio, S. Fancelli, and R. Fani. 1995. Identification of Azospirillum strains by restriction fragment length polymorphism of the 16S rDNA and the histidine operon. FEMS Microbiol. Lett. 127:85-91. [DOI] [PubMed] [Google Scholar]
- 5.Janse, J. D., M. P. Rossi, L. Angelucci, M. Scortichini, J. H. J. Derks, A. D. L. Akkermans, R. De Vrijer, and P. G. Psallidas. 1996. Reclassification of Pseudomonas syringae pv. avellanae as Pseudomonas avellanae (spec. nov.), the bacterium causing canker of hazelnut (Corylus avellana L.). Syst. Appl. Microbiol. 19:589-595. [Google Scholar]
- 6.King, E. O., M. K. Raney, and D. E. Ward. 1954. Two simple media for the demonstration of pyocianin and fluorescin. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 7.Laguerre, G., L. Rigottier-Gois, and P. Lemanceau. 1994. Fluorescent Pseudomonas species categorized by using polymerase chain reaction (PCR)/restriction fragment analysis of 16S rDNA. Mol. Ecol. 3:479-487. [DOI] [PubMed] [Google Scholar]
- 8.Lelliott, R. A., and D. E. Stead. 1987. Methods for the diagnosis of bacterial diseases of plants. Blackwell Scientific Publications for the British Society of Plant Pathology, Oxford, United Kingdom.
- 9.Louws, F. J., D. W. Fulbright, C. T. Stephens, and F. J. De Bruijn. 1994. Specific genomic fingerprinting of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl. Environ. Microbiol. 60:2285-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maynard Smith, J., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milgroom, M. G., and W. E. Fry. 1997. Contribution of population genetics to plant disease epidemiology and management. Adv. Bot. Res. 24:1-30. [Google Scholar]
- 12.Picard, C., F. Di Cello, M. Ventura, R. Fani, and A. Guckert. 2000. Frequency and biodiversity of 2,4-diacetylphloroglucinol-producing bacteria isolated from the maize rhizosphere at different stages of plant growth. Appl. Environ. Microbiol. 66:948-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Psallidas, P. G. 1987. The problem of bacterial canker of hazelnut in Greece caused by Pseudomonas syringae pv. avellanae. Bull. OEPP (Eur. Mediterran. Plant Prot. Org.)/EPPO 17:257-261.
- 14.Roos, I. L., Y. Alami, P. R. Harvey, K. W. Achouak, and M. H. Ryder. 2000. Genetic diversity and biological control activity of novel species of closely related pseudomonads isolated from wheat field soils in South Australia. Appl. Environ. Microbiol. 66:1609-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scortichini, M., and M. Lazzari. 1996. Systemic migration of Pseudomonas syringae pv. avellanae in twigs and young trees of hazelnut and symptom development. J. Phytopathol. 144:215-219. [Google Scholar]
- 16.Scortichini, M. 1998. Response of Corylus avellana germplasm to artificial inoculation with Pseudomonas avellanae (Psallidas) Janse et al. Agric. Mediterr. 128:153-156. [Google Scholar]
- 17.Scortichini, M., M. T. Dettori, U. Marchesi, M. A. Palombi, and M. P. Rossi. 1998. Differentiation of Pseudomonas avellanae strains from Greece and Italy by rep-PCR genomic fingerprinting. J. Phytopathol. 146:417-420. [Google Scholar]
- 18.Scortichini, M., U. Marchesi, M. P. Rossi, L. Angelucci, and M. T. Dettori. 2000. Rapid identification of Pseudomonas avellanae field isolates, causing hazelnut decline in central Italy, by repetitive PCR genomic fingerprinting. J. Phytopathol. 148:153-159. [Google Scholar]
- 19.Scortichini, M., U. Marchesi, M. T. Dettori, L. Angelucci, M. P. Rossi, and C. Morone. 2000. Genetic and pathogenic diversity of Pseudomonas avellanae srtrains isolated from Corylus avellana trees in north-west of Italy, and comparison with strains from other regions. Eur. J. Plant Pathol. 106:147-154. [Google Scholar]
- 20.Smith, J. J., L. C. Offord, M. Holderness, and G. S. Saddler. 1995. Genetic diversity of Burkholderia solanacearum (synonym, Pseudomonas solanacearum) race 3 in Kenya. Appl. Environ. Microbiol. 61:4263-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sorensen, K. N., K.-H. Kim, and J. Y. Takemoto. 1998. PCR detection of cyclic lipodepsinonapeptide-producing Pseudomonas syringae pv. syringae and similarity of strains. Appl. Environ. Microbiol. 64:226-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaneechoutte, M., R. Rossau, P. De Vos, M. Gillis, D. Janssens, N. Paepe, A. De Rouck, T. Fiers, G. Claeys, and K. Kersters. 1992. Rapid identification of bacteria of the Comamonadaceae with amplified ribosomal DNA-restriction analysis (ARDRA). FEMS Microbiol. Lett. 93:227-234. [DOI] [PubMed] [Google Scholar]
- 23.Vauterin, L., P. Yang, B. Hoste, M. Vancanneyt, E. L. Civerolo, J. Swings, and K. Kersters. 1991. Differentiation of Xanthomonas campestris pv. citri strains by sodium dodecyl sulfate-polyacrylamide gel electrophoresis of proteins, fatty acid analysis, and DNA-DNA hybridization. Int. J. Syst. Bacteriol. 41:535-542. [Google Scholar]
- 24.Yessad, S., C. Manceau, and J. Luisetti. 1992. A detached leaf assay to evaluate virulence and pathogenicity of strains of Pseudomonas syringae pv. syringae on pear. Plant Dis. 76:370-373. [Google Scholar]
- 25.Young, J. M. 1991. Pathogenicity and identification of the lilac pathogen Pseudomonas syringae pv. syringae van Hall. Ann. Appl. Biol. 118:283-298. [Google Scholar]
- 26.Young, J. M., D. S. Jones, and M. Gillis. 1996. Relationships between populations of Pseudomonas syringae pv. persicae determined by restriction fragment analysis. Plant Pathol. 45:350-357. [Google Scholar]