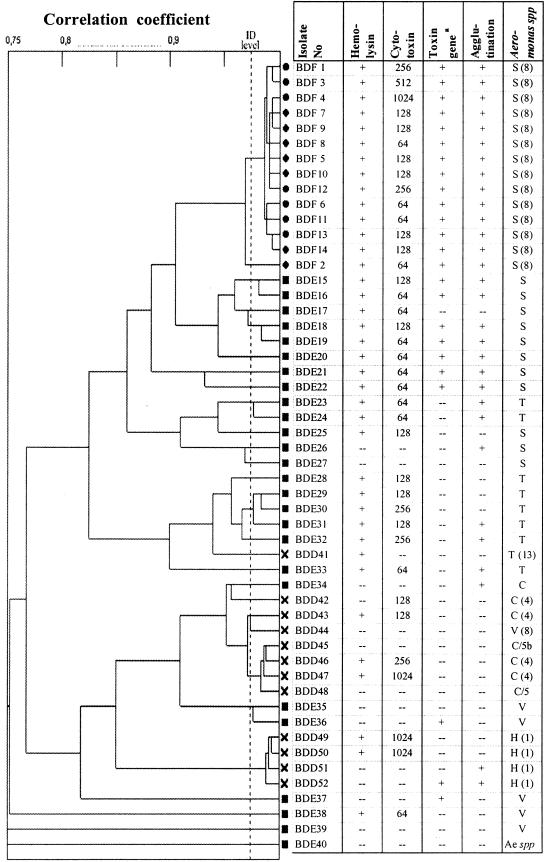
FIG. 1.
Dendrogram showing UPGMA clustering of PhP typing data obtained from 52 Aeromonas isolates. Circles indicate isolates from fish, squares indicate isolates from the environment, and crosses indicate isolates from humans with diarrhea. The results from assays of putative virulence properties are shown. The last column indicates the species codes. Some isolates were identified by FAME analysis, in which case the hybridization group (HG) is given, e.g., S(8) = A. veronii biovar sobria (hybridization group). Other isolates were identified by biochemical test: S, A. veronii biovar sobria; T, A. trota; C, A. caviae complex; H, A. hydrophila, V, A. veronii biovar veronii, etc. The column heading “toxin gene” refers to the cytolytic enterotoxin gene and/or extracellular hemolysin gene.